7M6S
 
 | |
5VPB
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-I crystal | | Descriptor: | CHLORIDE ION, Protein fosB, Transcription factor jun-D | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
8W3T
 
 | |
8VV5
 
 | |
7M6O
 
 | | Full length alpha1 Glycine receptor in presence of 0.1mM Glycine and 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
5VPI
 
 | | Crystal structure of human KRAS G12A mutant in complex with GTP | | Descriptor: | GTPase KRas, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Xu, S, Long, B, Boris, G, Ni, S, Kennedy, M.A. | | Deposit date: | 2017-05-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insight into the rearrangement of the switch I region in GTP-bound G12A K-Ras.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8VW6
 
 | | Crystal structure of SsuD | | Descriptor: | Alkanesulfonate monooxygenase | | Authors: | Caputo, A.T, Hu, M, Scott, C. | | Deposit date: | 2024-01-31 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystral structure of SsuD, a desulfonase protein from Rhodococcus jostii RHA
TBD
|
|
5VPO
 
 | | The 70S P-site ASL SufA6 complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hong, S, Sunita, S, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2017-05-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Mechanism of tRNA-mediated +1 ribosomal frameshifting.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7M6P
 
 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alphaZ1 | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
5VN5
 
 | | Crystal structure of LigY from Sphingobium sp. strain SYK-6 | | Descriptor: | 2,2',3-trihydroxy-3'-methoxy-5,5'-dicarboxybiphenyl meta-cleavage compound hydrolase, CHLORIDE ION, ZINC ION | | Authors: | Kuatsjah, E, Chan, A.C.K, Kobylarz, M.J, Murphy, M.E.P, Eltis, L.D. | | Deposit date: | 2017-04-28 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The bacterialmeta-cleavage hydrolase LigY belongs to the amidohydrolase superfamily, not to the alpha / beta-hydrolase superfamily.
J. Biol. Chem., 292, 2017
|
|
7LRK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-093 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{S})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
8VX3
 
 | | Structure of an exported small alarmone synthetase from Streptomyces albidoflavus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Ahmad, S, Tsang, K.K, Schiefer, V, Kim, Y, Whitney, J.C. | | Deposit date: | 2024-02-03 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a (p)ppApp synthetase from Streptomyces albidoflavus
To Be Published
|
|
5I8Q
 
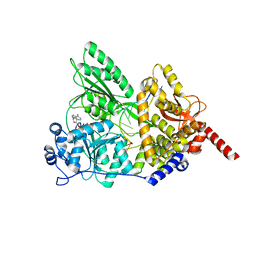 | | S. cerevisiae Prp43 in complex with RNA and ADPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP43, ... | | Authors: | He, Y, Nielsen, K.H, Andersen, G.R. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of the DEAH/RHA ATPase Prp43p bound to RNA implicates a pair of hairpins and motif Va in translocation along RNA.
RNA, 23, 2017
|
|
5VO1
 
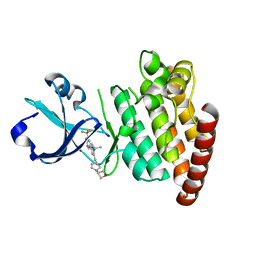 | |
8VNZ
 
 | |
7MAU
 
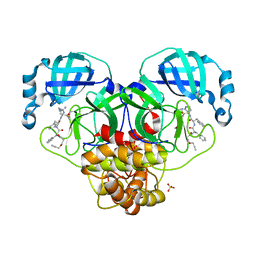 | |
8VXH
 
 | |
8VO9
 
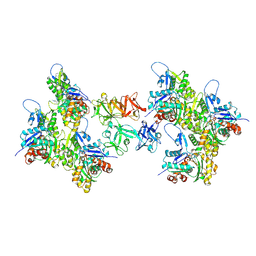 | | Cryo-EM structure of fascin crosslinked F-actin (Eigen_middle) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2024-01-14 | | Release date: | 2025-01-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Fascin structural plasticity mediates flexible actin bundle construction.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6W5H
 
 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5d | | Descriptor: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
8VVF
 
 | | Kappa opioid receptor:Galphai protein in complex with inverse agonist JDTic | | Descriptor: | (3R)-7-hydroxy-N-{(2S)-1-[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]-3-methylbutan-2-yl}-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gati, C, Motiwala, Z, Tyson, A.S, Styrpejko, D, Han, G.W, Khan, S, Ramos-Gonzalez, N, Shenvi, R, Majumdar, S. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanisms of inverse agonism via kappa-opioid receptor-G protein complexes.
Nat.Chem.Biol., 21, 2025
|
|
7LRO
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-01-105 | | Descriptor: | 1,2-ETHANEDIOL, 5-(azetidin-3-ylamino)-4-chloranyl-2-methyl-pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
8W2E
 
 | | SARS-CoV-2 M protein dimer in complex with JNJ-9676 and Fab-B | | Descriptor: | (6S,8R)-N-(3-cyanophenyl)-5-{4-[difluoro(phenyl)methyl]phenyl}-6-methyl-4-oxo-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazine-3-carboxamide, Fab B Heavy Chain, Fab B Light Chain, ... | | Authors: | Yin, Y, Van Damme, E. | | Deposit date: | 2024-02-20 | | Release date: | 2025-01-29 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | A small-molecule SARS-CoV-2 inhibitor targeting the membrane protein.
Nature, 640, 2025
|
|
7MAA
 
 | | HIV-1 Protease (I84V) in Complex with UMass10 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-ethylbutyl)amino]-1-benzyl-2-hydroxypropyl}carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with UMass10
To Be Published
|
|
5VD8
 
 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-099, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 6-{[3-chloro-4-(4-methylpiperazin-1-yl)phenyl]amino}-1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
7MAR
 
 | | Drug Resistant HIV-1 Protease (L10F, M46I, I47V, I50V, F53L, L63P, I72V, G73S, V82I, I85V) in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Drug Resistant HIV-1 Protease (L10F, M46I, I47V, I50V, F53L, L63P, I72V, G73S, V82I, I85V) in Complex with DRV
To Be Published
|
|
