9BDU
 
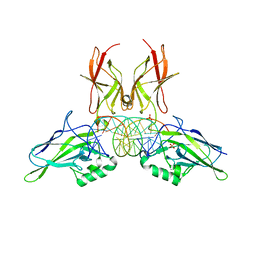 | | NF-kappaB RelA homo-dimer bound to AT-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BD3
 
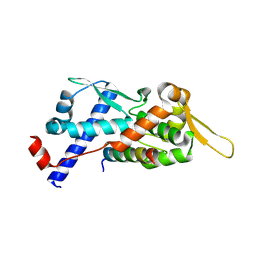 | | Structure of the MAGEA4 MHD-RAD18 R6BD Complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Melanoma antigen A 4 | | Authors: | Forker, K, Zhou, P. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of MAGEA4 MHD-RAD18 R6BD reveals a flipped binding mode compared to AlphaFold2 prediction.
Embo J., 2024
|
|
9BD0
 
 | |
9BCZ
 
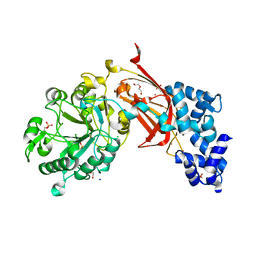 | | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Edwards, M.M, Dong, A, Theo-Emegano, N, Seitova, A, Loppnau, P, Leung, R, Li, H, Ilyassov, O, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-10 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine
To be published
|
|
9BCB
 
 | |
9BC4
 
 | | Transglutaminase 2 - Intermediate State | | Descriptor: | CALCIUM ION, GLYCEROL, HB-225 (gluten peptidomimetic TG2 inhibitor), ... | | Authors: | Sewa, A.S, Mathews, I.I, Khosla, C. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and mechanistic analysis of Ca 2+ -dependent regulation of transglutaminase 2 activity using a Ca 2+ -bound intermediate state.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BC3
 
 | | Transglutaminase 2 - Alternate state | | Descriptor: | CALCIUM ION, HB-225 (gluten peptidomimetic TG2 inhibitor), Protein-glutamine gamma-glutamyltransferase 2 | | Authors: | Mathews, I.I, Sewa, A.S, Khosla, C. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural and mechanistic analysis of Ca 2+ -dependent regulation of transglutaminase 2 activity using a Ca 2+ -bound intermediate state.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BC2
 
 | | Transglutaminase 2 - Open State | | Descriptor: | CHLORIDE ION, HB-225 (gluten peptidomimetic TG2 inhibitor), Protein-glutamine gamma-glutamyltransferase 2, ... | | Authors: | Mathews, I.I, Sewa, A, Khosla, C. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and mechanistic analysis of Ca 2+ -dependent regulation of transglutaminase 2 activity using a Ca 2+ -bound intermediate state.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BBM
 
 | |
9BBL
 
 | |
9BBI
 
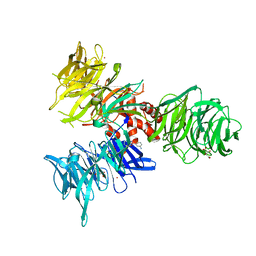 | | Co-crystal structure of human DDB1 bound to fragment UB028669 | | Descriptor: | 3-([1,3]oxazolo[4,5-b]pyridin-2-yl)aniline, DNA damage-binding protein 1, L(+)-TARTARIC ACID, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028669
To be published
|
|
9BBH
 
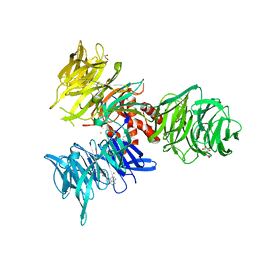 | | Co-crystal structure of human DDB1 bound to fragment UB028670 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole, DNA damage-binding protein 1, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028670
To be published
|
|
9BBG
 
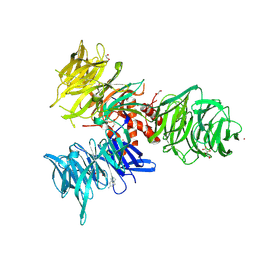 | | Co-crystal structure of human DDB1 bound to fragment UB028671 | | Descriptor: | 1,2-ETHANEDIOL, 1H-indol-6-amine, DNA damage-binding protein 1, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028671
To be published
|
|
9BBE
 
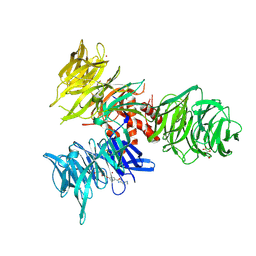 | | Co-crystal structure of human DDB1 bound to fragment UB028668 | | Descriptor: | 5-(4-methoxyphenyl)-3-[(3S)-pyrrolidin-3-yl]-1,2,4-oxadiazole, DNA damage-binding protein 1, L(+)-TARTARIC ACID, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028668
To be published
|
|
9BBB
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, cobicistat | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-04-05 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction of CYP3A4 with the inhibitor cobicistat: Structural and mechanistic insights and comparison with ritonavir
to be published
|
|
9BB7
 
 | |
9BB6
 
 | |
9BB5
 
 | |
9BB4
 
 | |
9BB3
 
 | |
9BB2
 
 | |
9BB1
 
 | |
9BAF
 
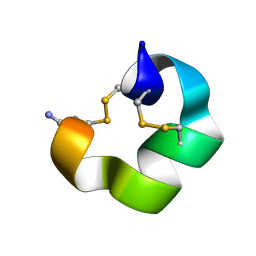 | | Solution NMR structure of conofurin-Delta | | Descriptor: | Alpha-conotoxin LvIA | | Authors: | Harvey, P.J, Craik, D.J, Hone, A.J, McIntosh, J.M. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Design, Synthesis, and Structure-Activity Relationships of Novel Peptide Derivatives of the Severe Acute Respiratory Syndrome-Coronavirus-2 Spike-Protein that Potently Inhibit Nicotinic Acetylcholine Receptors.
J.Med.Chem., 67, 2024
|
|
9BA5
 
 | |
9BA4
 
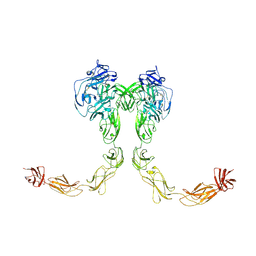 | | Full-length cross-linked Contactin 2 (CNTN2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2 | | Authors: | Liu, J.L, Fan, S.F, Ren, G.R, Rudenko, G.R. | | Deposit date: | 2024-04-03 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of Contactin 2 homophilic interaction
To Be Published
|
|
