1FAS
 
 | | 1.9 ANGSTROM RESOLUTION STRUCTURE OF FASCICULIN 1, AN ANTI-ACETYLCHOLINESTERASE TOXIN FROM GREEN MAMBA SNAKE VENOM | | Descriptor: | FASCICULIN 1 | | Authors: | Le Du, M.H, Marchot, P, Bougis, P.E, Fontecilla-Camps, J.C. | | Deposit date: | 1992-08-07 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.9-A resolution structure of fasciculin 1, an anti-acetylcholinesterase toxin from green mamba snake venom.
J.Biol.Chem., 267, 1992
|
|
3ZGZ
 
 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and toxic moiety from agrocin 84 (TM84) in aminoacylation-like conformation | | Descriptor: | LEUCINE--TRNA LIGASE, MAGNESIUM ION, TRNA-LEU UAA ISOACCEPTOR, ... | | Authors: | Chopra, S, Palencia, A, Virus, C, Tripathy, A, Temple, B.R, Velazquez-Campoy, A, Cusack, S, Reader, J.S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Plant Tumour Biocontrol Agent Employs a tRNA-Dependent Mechanism to Inhibit Leucyl-tRNA Synthetase
Nat.Commun., 4, 2013
|
|
1FRV
 
 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF NI-FE HYDROGENASE | | Descriptor: | FE3-S4 CLUSTER, HYDRATED FE, HYDROGENASE, ... | | Authors: | Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1996-03-28 | | Release date: | 1996-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas.
Nature, 373, 1995
|
|
3UA8
 
 | | Crystal Structure Analysis of a 6-Amino Quinazolinedione Sulfonamide bound to human GluR2 | | Descriptor: | Glutamate receptor 2, N-methyl-1-{3-[(methylsulfonyl)amino]-2,4-dioxo-7-(trifluoromethyl)-1,2,3,4-tetrahydroquinazolin-6-yl}-1H-imidazole-4-carboxamide | | Authors: | Kallen, J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 6-Amino quinazolinedione sulfonamides as orally active competitive AMPA receptor antagonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3KZM
 
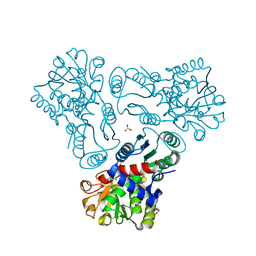 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate | | Descriptor: | GLYCEROL, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
3KZO
 
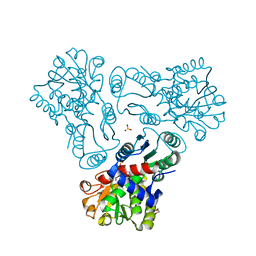 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate and N-acetyl-L-norvaline | | Descriptor: | GLYCEROL, N-ACETYL-L-NORVALINE, N-acetylornithine carbamoyltransferase, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
3KZN
 
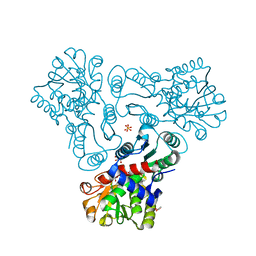 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with N-acetyl-L-ornirthine | | Descriptor: | GLYCEROL, N-acetylornithine carbamoyltransferase, N~2~-ACETYL-L-ORNITHINE, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
4JY8
 
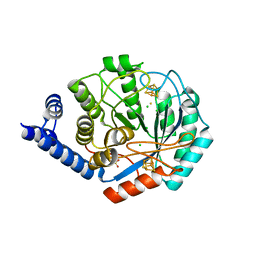 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | Descriptor: | CHLORIDE ION, FEFE-HYDROGENASE MATURASE, HYDROSULFURIC ACID, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3CIW
 
 | | X-RAY structure of the [FeFe]-hydrogenase maturase HydE from thermotoga maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, FeFe-Hydrogenase maturase, ... | | Authors: | Nicolet, Y, Ruback, J.K, Posewitz, M.C, Amara, P, Mathevon, C, Atta, M, Fontecave, M, Fontecilla-Camps, J.C. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray Structure of the [FeFe]-Hydrogenase Maturase HydE from Thermotoga maritima
J.Biol.Chem., 283, 2008
|
|
3SM2
 
 | | The crystal structure of XMRV protease complexed with Amprenavir | | Descriptor: | gag-pro-pol polyprotein, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.
Febs J., 278, 2011
|
|
2AO7
 
 | | Adam10 Disintegrin and cysteine- rich domain | | Descriptor: | ADAM 10, SULFATE ION | | Authors: | Janes, P.W, Saha, N, Barton, W.A, Kolev, M.V, Wimmer-Kleikamp, S.H, Nievergall, E, Blobel, C.P, Himanen, J.-P, Lackmann, M, Nikolov, D.B. | | Deposit date: | 2005-08-12 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans.
Cell(Cambridge,Mass.), 123, 2005
|
|
2XHD
 
 | | Crystal structure of N-((2S)-5-(6-fluoro-3-pyridinyl)-2,3-dihydro-1H- inden-2-yl)-2-propanesulfonamide in complex with the ligand binding domain of the human GluA2 receptor | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N-[(2S)-5-(6-FLUORO-3-PYRIDINYL)-2,3-DIHYDRO-1H-INDEN-2-YL]-2-PROPANESULFONAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Andreotti, D, Ballantine, S, Bax, B.D, Harris, A.J, Harker, A.J, Lund, J, Melarange, R, Mingardi, A, Mookherjee, C, Mosley, J, Neve, M, Oliosi, B, Profeta, R, Smith, K.J, Smith, P.W, Spada, S, Thewlis, K.M, Yusaf, S.P. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of N-[(2S)-5-(6-Fluoro-3-Pyridinyl)-2,3-Dihydro-1H-Inden-2-Yl]-2-Propanesulfonamide, a Novel Clinical Ampa Receptor Positive Modulator.
J.Med.Chem., 53, 2010
|
|
1CLQ
 
 | |
4JJ0
 
 | | Crystal structure of MamP | | Descriptor: | GLYCEROL, HEME C, MamP | | Authors: | Siponen, M, Pignol, D, Arnoux, P. | | Deposit date: | 2013-03-07 | | Release date: | 2013-10-09 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into magnetochrome-mediated magnetite biomineralization.
Nature, 502, 2013
|
|
4JY9
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | Descriptor: | CHAPSO, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYF
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | CARBONATE ION, CHAPSO, CHLORIDE ION, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYE
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | BROMIDE ION, Biotin synthetase, putative, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYD
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, BROMIDE ION, CHAPSO, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1DQB
 
 | |
4JJ3
 
 | |
2XWE
 
 | | X-RAY STRUCTURE OF ACID-BETA-GLUCOSIDASE WITH 5N,6S-(N'-(N-OCTYL)IMINO)-6-THIONOJIRIMYCIN IN THE ACTIVE SITE | | Descriptor: | (3Z,5S,6R,7S,8R,8aS)-3-(octylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-5,6,7,8-tetrol, GLUCOSYLCERAMIDASE, PHOSPHATE ION, ... | | Authors: | Brumshtein, B, Aguilar-Moncayo, M, Benito, J.M, Ortiz Mellet, C, Garcia Fernandez, J.M, Silman, I, Shaaltiel, Y, Sussman, J.L, Futerman, A.H. | | Deposit date: | 2010-11-02 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Cyclodextrin-Mediated Crystallization of Acid Beta-Glucosidase in Complex with Amphiphilic Bicyclic Nojirimycin Analogues.
Org.Biomol.Chem., 9, 2011
|
|
4P3X
 
 | | Structure of the Fe4S4 quinolinate synthase NadA from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Cherrier, M.V, Chan, A, Darnault, C, Reichmann, D, Amara, P, Ollagnier de Choudens, S, Fontecilla-Camps, J.C. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of Fe4S4 quinolinate synthase unravels an enzymatic dehydration mechanism that uses tyrosine and a hydrolase-type triad.
J.Am.Chem.Soc., 136, 2014
|
|
1E08
 
 | | Structural model of the [Fe]-Hydrogenase/cytochrome c553 complex combining NMR and soft-docking | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Morelli, X, Czjzek, M, Hatchikian, C.E, Bornet, O, Fontecilla-Camps, J.C, Palma, N.P, Moura, J.J.G, Guerlesquin, F. | | Deposit date: | 2000-03-13 | | Release date: | 2000-08-25 | | Last modified: | 2019-11-27 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Model of the Fe-Hydrogenase/Cytochrome C553 Complex Combining Transverse Relaxation-Optimized Spectroscopy Experiments and Soft Docking Calculations.
J.Biol.Chem., 275, 2000
|
|
2FQ8
 
 | |
2XEV
 
 | |
