5DS1
 
 | | Core domain of the class II small heat-shock protein HSP 17.7 from Pisum sativum | | Descriptor: | 17.1 kDa class II heat shock protein | | Authors: | Hochberg, G.K.A, Laganoswky, A, Allison, T.A, Shepherd, D.A, Benesch, J.L.P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural principles that enable oligomeric small heat-shock protein paralogs to evolve distinct functions.
Science, 359, 2018
|
|
3DBG
 
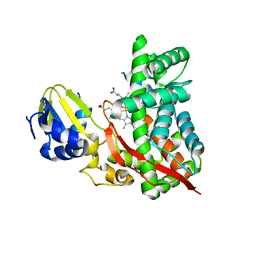 | |
6UNK
 
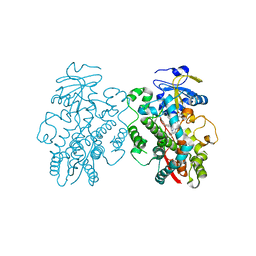 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(naphthalen-1-yl)-3-{[(2R)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-10-12 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
5E5E
 
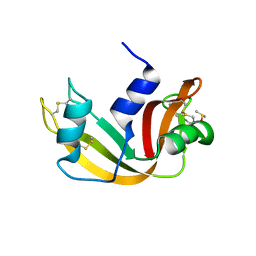 | |
5DT4
 
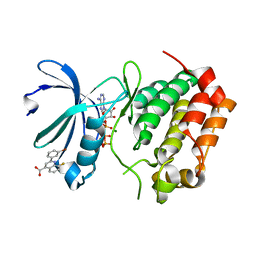 | | Aurora A Kinase in Complex with AA35 and ATP in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
6H1S
 
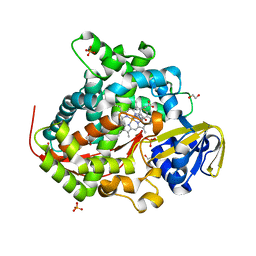 | | Structure of the BM3 heme domain in complex with fluconazole | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
6GOM
 
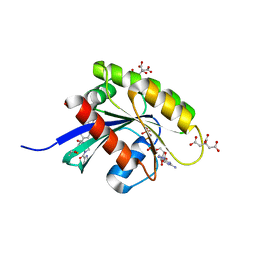 | | KRAS-169 Q61H GPPNHP + PPIN-1 | | Descriptor: | (6~{S})-1-(1~{H}-imidazol-4-ylcarbonyl)-6-[(4-phenylphenyl)methyl]-4-propyl-1,4-diazepan-5-one, CITRIC ACID, GTPase KRas, ... | | Authors: | Cruz-Migoni, A, Canning, P, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2018-06-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3DCV
 
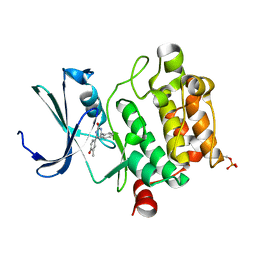 | | Crystal structure of human Pim1 kinase complexed with 4-(4-hydroxy-3-methyl-phenyl)-6-phenylpyrimidin-2(1H)-one | | Descriptor: | 4-(4-hydroxy-3-methylphenyl)-6-phenylpyrimidin-2(5H)-one, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Bellamacina, C.R, Shafer, C.M, Lindvall, M, Gesner, T.G, Yabannavar, A, Weiping, J, Song, L, Walter, A. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 4-(1H-indazol-5-yl)-6-phenylpyrimidin-2(1H)-one analogs as potent CDC7 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5DU3
 
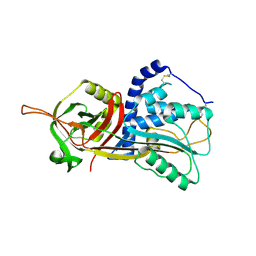 | | Active form of human C1-inhibitor | | Descriptor: | Plasma protease C1 inhibitor | | Authors: | Pannu, N.S, Dijk, M, Holkers, J, Voskamp, P, Giannetti, B.M, Waterreus, W.J, van Veen, H.A. | | Deposit date: | 2015-09-18 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How Dextran Sulfate Affects C1-inhibitor Activity: A Model for Polysaccharide Potentiation.
Structure, 24, 2016
|
|
6GQE
 
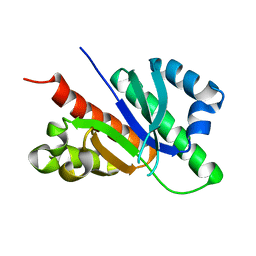 | |
6H46
 
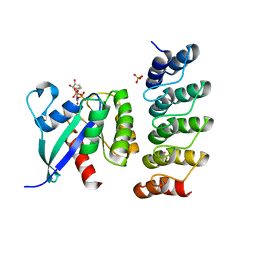 | | Human KRAS in complex with darpin K13 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Debreczeni, J.E, Bery, N, Legg, S, Breed, J, Embrey, K, Stubbs, C, Kolasinska-Zwierz, P, Barrett, N, Marwood, R, Watson, J, Tart, J, Overman, R, Miller, A, Phillips, C, Minter, R, Rabbitts, T.H. | | Deposit date: | 2018-07-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | KRAS-specific inhibition using a DARPin binding to a site in the allosteric lobe.
Nat Commun, 10, 2019
|
|
3DE6
 
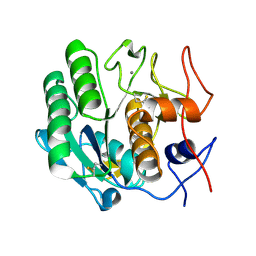 | |
6TZE
 
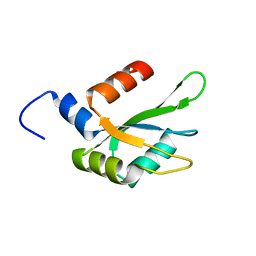 | | Human CstF-64 RRM mutant - D50A | | Descriptor: | Cleavage stimulation factor subunit 2 | | Authors: | Latham, M.P, Masoumzadeh, E. | | Deposit date: | 2019-08-12 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A missense mutation in the CSTF2 gene that impairs the function of the RNA recognition motif and causes defects in 3' end processing is associated with intellectual disability in humans.
Nucleic Acids Res., 48, 2020
|
|
8AEH
 
 | |
6UNH
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-(1H-indol-3-yl)-3-{[(2S)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
8AE8
 
 | |
3DFC
 
 | |
3DA6
 
 | | Crystal Structure of human JNK3 complexed with N-(3-methyl-4-(3-(2-(methylamino)pyrimidin-4-yl)pyridin-2-yloxy)naphthalen-1-yl)-1H-benzo[d]imidazol-2-amine | | Descriptor: | Mitogen-activated protein kinase 10, N-[3-methyl-4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-1H-benzimidazol-2-amine | | Authors: | Cee, V.J, Cheng, A.C, Romero, K, Bellon, S, Mohr, C, Whittington, D.A, Bready, J, Caenepeel, S, Coxon, A, Deak, H.L, Hodous, B.L, Kim, J.L, Lin, J, Nguyen, H, Olivieri, P.R, Patel, V.F, Wang, L, Hughes, P, Geuns-Meyer, S. | | Deposit date: | 2008-05-28 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5EAG
 
 | | Saccharomyces cerevisiae CYP51 complexed with the plant pathogen inhibitor Prochloraz | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, Prochloraz | | Authors: | Tyndall, J.D.A, Sabherwal, M, Sagatova, A.A, Keniya, M.V, Wilson, R.K, Woods, M.V, Monk, B.C. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Elucidation of Yeast Lanosterol 14 alpha-Demethylase in Complex with Agrochemical Antifungals.
PLoS ONE, 11, 2016
|
|
3DAJ
 
 | |
6U31
 
 | | The crystal structure of 4-(1H-imidazol-1-yl)benzoate-bound CYP199A4 | | Descriptor: | 4-(1H-imidazol-1-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | To Be, or Not to Be, an Inhibitor: A Comparison of Azole Interactions with and Oxidation by a Cytochrome P450 Enzyme.
Inorg.Chem., 61, 2022
|
|
3DGI
 
 | | Crystal structure of F87A/T268A mutant of CYP BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Le Trong, I, Katayama, J.H, Totah, R.A, Stenkamp, R.E, Fox, E.P. | | Deposit date: | 2008-06-13 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Homolytic versus heterolytic dioxygen bond cleavage in cytochrome P450 BM3.
To be Published
|
|
3DH6
 
 | | Crystal structure of bovine pancreatic ribonuclease A variant (V47A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
6HME
 
 | | LOW-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA; CSNK2A1 gene product) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR THN27 | | Descriptor: | 1,2-ETHANEDIOL, 5-propan-2-yl-4-prop-2-enoxy-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
6U1Y
 
 | | bcs1 AAA domain | | Descriptor: | MAGNESIUM ION, Mitochondrial chaperone BCS1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Tang, W.K, Xia, D. | | Deposit date: | 2019-08-17 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
