8TNO
 
 | |
8TNM
 
 | |
4O5T
 
 | | Crystal structure of Diels-Alderase CE20 in complex with a product analog | | Descriptor: | 4-{[2-(phosphonooxy)ethyl]carbamoyl}benzyl [(1R,6S)-6-(dimethylcarbamoyl)cyclohex-2-en-1-yl]carbamate, Diisopropyl-fluorophosphatase | | Authors: | Beck, T, Preiswerk, N, Mayer, C, Hilvert, D. | | Deposit date: | 2013-12-20 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Impact of scaffold rigidity on the design and evolution of an artificial Diels-Alderase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O60
 
 | | Structure of ankyrin repeat protein | | Descriptor: | ANK-N5C-317 | | Authors: | Ethayathulla, A.S, Guan, L. | | Deposit date: | 2013-12-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A transcription blocker isolated from a designed repeat protein combinatorial library by in vivo functional screen.
Sci Rep, 5, 2015
|
|
4O5S
 
 | |
6TJE
 
 | |
3QHT
 
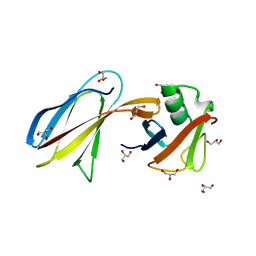 | | Crystal Structure of the Monobody ySMB-1 bound to yeast SUMO | | Descriptor: | GLYCEROL, Monobody ySMB-1, Ubiquitin-like protein SMT3 | | Authors: | Koide, S, Gilbreth, R.N. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6TJH
 
 | | Crystal structure of the computationally designed Cake9 protein | | Descriptor: | Cake9, GLYCEROL, SULFATE ION | | Authors: | Mylemans, B, Laier, I, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TJC
 
 | | Crystal structure of the computationally designed Cake3 protein | | Descriptor: | Cake3, GLYCEROL, PHOSPHATE ION | | Authors: | Laier, I, Mylemans, B, Voet, A.R.D, Noguchi, H. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TJG
 
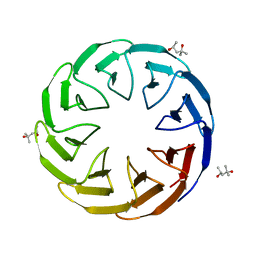 | | Crystal structure of the computationally designed Cake8 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake8 | | Authors: | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
8SJH
 
 | |
8SJF
 
 | |
8SJG
 
 | |
8SJI
 
 | |
4NW9
 
 | |
6V58
 
 | |
6V5G
 
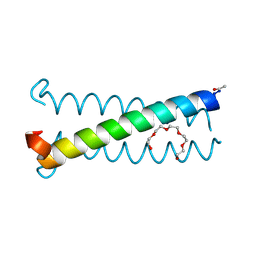 | | Coiled-coil Trimer with Ala:Leu:Lys Triad | | Descriptor: | Coiled-coil Trimer with Ala:Leu:Lys Triad, PENTAETHYLENE GLYCOL | | Authors: | Smith, M.S, Stern, K.L, Billings, W.M, Price, J.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Context-Dependent Stabilizing Interactions among Solvent-Exposed Residues along the Surface of a Trimeric Helix Bundle.
Biochemistry, 59, 2020
|
|
4P4V
 
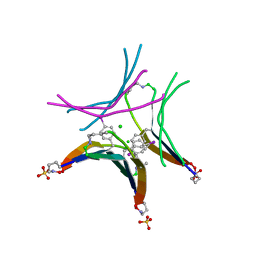 | |
4P4Z
 
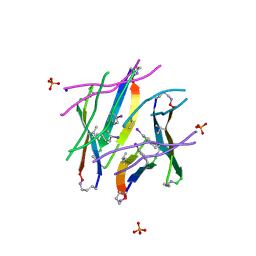 | |
6V57
 
 | |
6V5I
 
 | | Coiled-coil Trimer with Ala:Leu:Ala Triad | | Descriptor: | Coiled-coil Trimer with Ala:Leu:Ala Triad, PENTAETHYLENE GLYCOL | | Authors: | Smith, M.S, Stern, K.L, Billings, W.M, Price, J.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-Dependent Stabilizing Interactions among Solvent-Exposed Residues along the Surface of a Trimeric Helix Bundle.
Biochemistry, 59, 2020
|
|
6V5J
 
 | |
4QFV
 
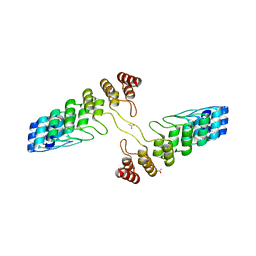 | |
6TJD
 
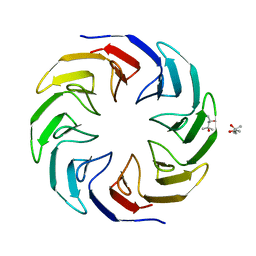 | | Crystal structure of the computationally designed Cake4 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake4 | | Authors: | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
4PJR
 
 | | Crystal structure of designed cPPR-NRE protein | | Descriptor: | MAGNESIUM ION, Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
