1AX7
 
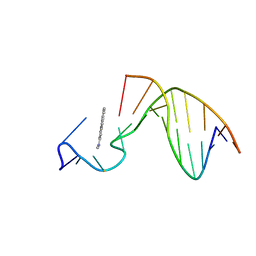 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED AT A TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(AAC-[AF]G-CTACCATCC)D(GGATGGTAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-stacked conformer of the syn [AF]-C8-dG adduct positioned at a template-primer junction.
Biochemistry, 36, 1997
|
|
1BCB
 
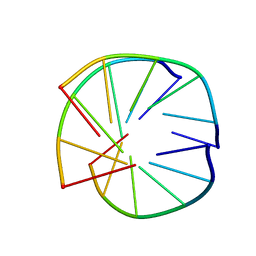 | | INTRAMOLECULAR TRIPLEX, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*T)-3') | | Authors: | Asensio, J.L, Brown, T, Lane, A.N. | | Deposit date: | 1998-04-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of intramolecular DNA triple helices containing adjacent and non-adjacent CG.C+ triplets.
Nucleic Acids Res., 26, 1998
|
|
1AFF
 
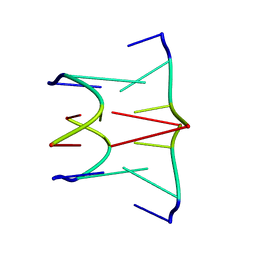 | | DNA QUADRUPLEX CONTAINING GGGG TETRADS AND (T.A).A TRIADS, NMR, 8 STRUCTURES | | Descriptor: | QUADRUPLEX DNA (5'-D(TP*AP*GP*G)-3') | | Authors: | Kettani, A, Bouaziz, S, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1997-03-06 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads.
Nat.Struct.Biol., 4, 1997
|
|
1AP7
 
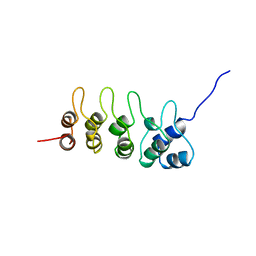 | | P19-INK4D FROM MOUSE, NMR, 20 STRUCTURES | | Descriptor: | P19-INK4D | | Authors: | Archer, S.J, Luh, F.Y, Domaille, P.J, Smith, B.O, Laue, E.D. | | Deposit date: | 1997-07-25 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the cyclin-dependent kinase inhibitor p19Ink4d.
Nature, 389, 1997
|
|
6I2O
 
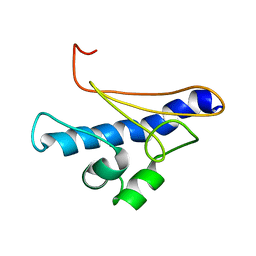 | |
1GYA
 
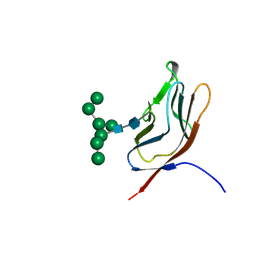 | | N-GLYCAN AND POLYPEPTIDE NMR SOLUTION STRUCTURES OF THE ADHESION DOMAIN OF HUMAN CD2 | | Descriptor: | HUMAN CD2, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wyss, D.F, Choi, J.S, Wagner, G. | | Deposit date: | 1995-05-26 | | Release date: | 1996-11-08 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Conformation and function of the N-linked glycan in the adhesion domain of human CD2.
Science, 269, 1995
|
|
1GZA
 
 | | PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of iodide to Arthromyces ramosus peroxidase investigated with X-ray crystallographic analysis, 1H and 127I NMR spectroscopy, and steady-state kinetics.
J.Biol.Chem., 272, 1997
|
|
6I1B
 
 | |
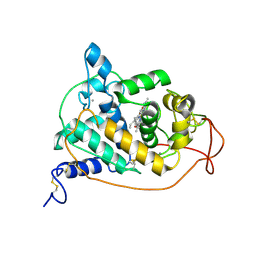
1JV4
 
 | | Crystal structure of recombinant major mouse urinary protein (rmup) at 1.75 A resolution | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, Major urinary protein 2 | | Authors: | Kuser, P.R, Franzoni, L, Ferrari, E, Spisni, A, Polikarpov, I. | | Deposit date: | 2001-08-28 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray structure of a recombinant major urinary protein at 1.75 A resolution. A comparative study of X-ray and NMR-derived structures.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5VKV
 
 | |
5ZCZ
 
 | | Solution structure of the T. Thermophilus HB8 TTHA1718 protein in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Heavy metal binding protein | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5ZD0
 
 | | Solution structure of human ubiquitin with three alanine mutations in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | ubiquitin | | Authors: | Tanaka, T, Ikeya, T, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6AX2
 
 | |
6B9K
 
 | |
6BHN
 
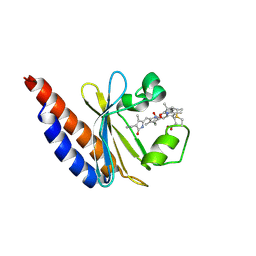 | | Red Light-Absorbing State of NpR6012g4, a Red/Green Cyanobacteriochrome | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with phytochrome sensor, PHYCOCYANOBILIN | | Authors: | Yu, Q, Lim, S, Rockwell, N.C, Martin, S.S, Lagarias, J.C, Ames, J.B. | | Deposit date: | 2017-10-31 | | Release date: | 2018-04-18 | | Last modified: | 2019-12-04 | | Method: | SOLUTION NMR | | Cite: | Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BGH
 
 | |
6BHO
 
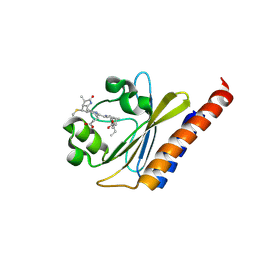 | | Green Light-Absorbing State of NpR6012g4, a Red/Green Cyanobacteriochrome | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with phytochrome sensor, PHYCOCYANOBILIN | | Authors: | Lim, S, Yu, Q, Rockwell, N.C, Martin, S.S, Lagarias, J.C, Ames, J.B. | | Deposit date: | 2017-10-31 | | Release date: | 2018-04-18 | | Last modified: | 2019-12-04 | | Method: | SOLUTION NMR | | Cite: | Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1LCC
 
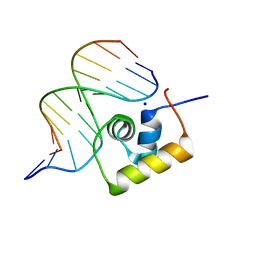 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
1LCD
 
 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
6BGG
 
 | | Solution NMR structures of the BRD3 ET domain in complex with a CHD4 peptide | | Descriptor: | Bromodomain-containing protein 3, CHD4 | | Authors: | Wai, D.C.C, Szyszka, T.N, Campbell, A.E, Kwong, C, Wilkinson-White, L, Silva, A.P.G, Low, J.K.K, Kwan, A.H, Gamsjaeger, R, Lu, B, Vakoc, C.R, Blobel, G.A, Mackay, J.P. | | Deposit date: | 2017-10-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.
J. Biol. Chem., 293, 2018
|
|
6C0A
 
 | | Actinin-1 EF-Hand bound to the Cav1.2 IQ Motif | | Descriptor: | Alpha-actinin-1, Voltage-dependent L-type calcium channel subunit alpha-1C | | Authors: | Turner, M.L, Ames, J.B. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Localization and Activation of Neuronal L-type Ca2+ Channels by a-Actinin1 and Calmodulin
To be Published
|
|
3GUZ
 
 | | Structural and substrate-binding studies of pantothenate synthenate (PS)provide insights into homotropic inhibition by pantoate in PS's | | Descriptor: | PANTOATE, Pantothenate synthetase | | Authors: | Chakrabarti, K.S, Thakur, K.G, Gopal, B, Sarma, S.P. | | Deposit date: | 2009-03-30 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray crystallographic and NMR studies of pantothenate synthetase provide insights into the mechanism of homotropic inhibition by pantoate
Febs J., 277, 2010
|
|
3EZB
 
 | | COMPLEX OF THE AMINO TERMINAL DOMAIN OF ENZYME I AND THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (PHOSPHOCARRIER PROTEIN HPR), PROTEIN (PHOSPHOTRANSFER SYSTEM, ENZYME I) | | Authors: | Clore, G.M, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1998-11-03 | | Release date: | 1999-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr.
Nat.Struct.Biol., 6, 1999
|
|
1AQA
 
 | | SOLUTION STRUCTURE OF REDUCED MICROSOMAL RAT CYTOCHROME B5, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Ferroni, F, Rosato, A. | | Deposit date: | 1997-07-28 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced microsomal rat cytochrome b5.
Eur.J.Biochem., 249, 1997
|
|
1AZ6
 
 | |
