4MBA
 
 | | APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | IMIDAZOLE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1989-02-22 | | Release date: | 1990-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution.
J.Mol.Biol., 205, 1989
|
|
2CBH
 
 | |
2HIR
 
 | |
2CA2
 
 | | CRYSTALLOGRAPHIC STUDIES OF INHIBITOR BINDING SITES IN HUMAN CARBONIC ANHYDRASE II. A PENTACOORDINATED BINDING OF THE SCN-ION TO THE ZINC AT HIGH P*H | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, THIOCYANATE ION, ... | | Authors: | Eriksson, A.E, Kylsten, P.M, Jones, T.A, Liljas, A. | | Deposit date: | 1989-02-06 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies of inhibitor binding sites in human carbonic anhydrase II: a pentacoordinated binding of the SCN- ion to the zinc at high pH.
Proteins, 4, 1988
|
|
1L26
 
 | |
1TLD
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-TRYPSIN AT 1.5 ANGSTROMS RESOLUTION IN A CRYSTAL FORM WITH LOW MOLECULAR PACKING DENSITY. ACTIVE SITE GEOMETRY, ION PAIRS AND SOLVENT STRUCTURE | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, SULFATE ION | | Authors: | Bartunik, H.D, Summers, L.J, Bartsch, H.H. | | Deposit date: | 1989-07-24 | | Release date: | 1990-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of bovine beta-trypsin at 1.5 A resolution in a crystal form with low molecular packing density. Active site geometry, ion pairs and solvent structure.
J.Mol.Biol., 210, 1989
|
|
1R08
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(2,6-DICHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PENTYL)-3-(HYDROXYETHYL OXYMETHYLENEOXYMETHYL) ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
3P2P
 
 | | ENHANCED ACTIVITY AND ALTERED SPECIFICITY OF PHOSPHOLIPASE A2 BY DELETION OF A SURFACE LOOP | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Dijkstra, B.W, Thunnissen, M.M.G.M, Kalk, K.H, Drenth, J. | | Deposit date: | 1989-11-29 | | Release date: | 1990-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhanced activity and altered specificity of phospholipase A2 by deletion of a surface loop.
Science, 244, 1989
|
|
1MBA
 
 | | APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1989-02-22 | | Release date: | 1990-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution.
J.Mol.Biol., 205, 1989
|
|
1I1B
 
 | |
3CA2
 
 | | CRYSTALLOGRAPHIC STUDIES OF INHIBITOR BINDING SITES IN HUMAN CARBONIC ANHYDRASE II. A PENTACOORDINATED BINDING OF THE SCN-ION TO THE ZINC AT HIGH P*H | | Descriptor: | 3-MERCURI-4-AMINOBENZENESULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Eriksson, A.E, Jones, T.A, Liljas, A. | | Deposit date: | 1989-10-02 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of inhibitor binding sites in human carbonic anhydrase II: a pentacoordinated binding of the SCN- ion to the zinc at high pH.
Proteins, 4, 1988
|
|
1RMU
 
 | | THREE-DIMENSIONAL STRUCTURES OF DRUG-RESISTANT MUTANTS OF HUMAN RHINOVIRUS 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Krishnaswamy, S, Kremer, M.J, Oliveira, M.A, Rossmann, M.G, Heinz, B.A, Rueckert, R.R, Dutko, F.J, Mckinlay, M.A. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of drug-resistant mutants of human rhinovirus 14.
J.Mol.Biol., 207, 1989
|
|
2R04
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2R07
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(6-CHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2R06
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2MEV
 
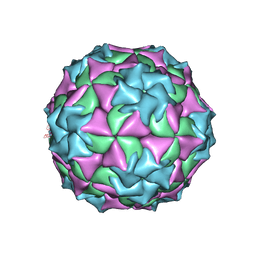 | | STRUCTURAL REFINEMENT AND ANALYSIS OF MENGO VIRUS | | Descriptor: | MENGO VIRUS COAT PROTEIN (SUBUNIT VP1), MENGO VIRUS COAT PROTEIN (SUBUNIT VP2), MENGO VIRUS COAT PROTEIN (SUBUNIT VP3), ... | | Authors: | Rossmann, M.G. | | Deposit date: | 1989-04-21 | | Release date: | 1990-01-15 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural refinement and analysis of Mengo virus.
J.Mol.Biol., 211, 1990
|
|
1P01
 
 | |
5XIA
 
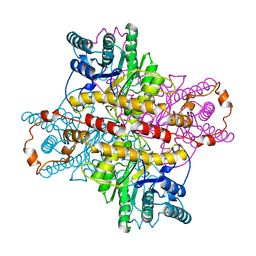 | | STRUCTURES OF D-XYLOSE ISOMERASE FROM ARTHROBACTER STRAIN B3728 CONTAINING THE INHIBITORS XYLITOL AND D-SORBITOL AT 2.5 ANGSTROMS AND 2.3 ANGSTROMS RESOLUTION, RESPECTIVELY | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Henrick, K, Collyer, C.A, Blow, D.M. | | Deposit date: | 1989-07-05 | | Release date: | 1990-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of D-xylose isomerase from Arthrobacter strain B3728 containing the inhibitors xylitol and D-sorbitol at 2.5 A and 2.3 A resolution, respectively.
J.Mol.Biol., 208, 1989
|
|
4XIA
 
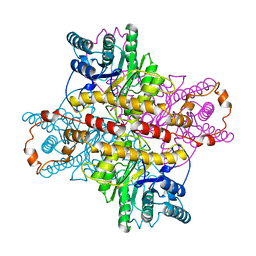 | | STRUCTURES OF D-XYLOSE ISOMERASE FROM ARTHROBACTER STRAIN B3728 CONTAINING THE INHIBITORS XYLITOL AND D-SORBITOL AT 2.5 ANGSTROMS AND 2.3 ANGSTROMS RESOLUTION, RESPECTIVELY | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, sorbitol | | Authors: | Henrick, K, Collyer, C.A, Blow, D.M. | | Deposit date: | 1989-07-05 | | Release date: | 1990-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-xylose isomerase from Arthrobacter strain B3728 containing the inhibitors xylitol and D-sorbitol at 2.5 A and 2.3 A resolution, respectively.
J.Mol.Biol., 208, 1989
|
|
5EBX
 
 | |
3CPP
 
 | |
1DN9
 
 | |
1P05
 
 | |
2I1B
 
 | |
5ANA
 
 | |
