7DVQ
 
 | | Cryo-EM Structure of the Activated Human Minor Spliceosome (minor Bact Complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'-O-[(S)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(methoxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]guanosine, Armadillo repeat-containing protein 7, ... | | Authors: | Bai, R, Wan, R, Wang, L, Xu, K, Zhang, Q, Lei, J, Shi, Y. | | Deposit date: | 2021-01-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the activated human minor spliceosome.
Science, 371, 2021
|
|
7EVN
 
 | | The cryo-EM structure of the DDX42-SF3b complex | | Descriptor: | ATP-dependent RNA helicase DDX42, PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome
Nat.Struct.Mol.Biol., 2024
|
|
7EVO
 
 | | The cryo-EM structure of the human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, RNA helicase, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome.
Nat.Struct.Mol.Biol., 2024
|
|
7FSE
 
 | | Crystal Structure of T. maritima reverse gyrase with a minimal latch | | Descriptor: | CHLORIDE ION, DODECAETHYLENE GLYCOL, Reverse gyrase, ... | | Authors: | Rasche, R, Kummel, D, Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2023-01-04 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7FSF
 
 | | CRYSTAL STRUCTURE OF T. MARITIMA REVERSE GYRASE ACTIVE SITE VARIANT Y851F | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rasche, R, Kummel, D, Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2023-01-04 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7GQS
 
 | | Crystal Structure of Werner helicase fragment 517-945 in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ... | | Authors: | Classen, M, Benz, J, Brugger, D, Rudolph, M.G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase.
Nature, 629, 2024
|
|
7GQT
 
 | | Crystal Structure of Werner helicase fragment 517-945 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, MAGNESIUM ION, ... | | Authors: | Classen, M, Benz, J, Brugger, D, Rudolph, M.G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase.
Nature, 629, 2024
|
|
7GQU
 
 | | Crystal Structure of Werner helicase fragment 517-945 in covalent complex with N-[(E,1S)-1-cyclopropyl-3-methylsulfonylprop-2-enyl]-2-(1,1-difluoroethyl)-4-phenoxypyrimidine-5-carboxamide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, GLYCEROL, ... | | Authors: | Classen, M, Benz, J, Brugger, D, Tagliente, O, Rudolph, M.G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase.
Nature, 629, 2024
|
|
7JL1
 
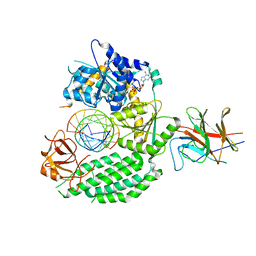 | | Cryo-EM structure of RIG-I:dsRNA in complex with RIPLET PrySpry domain (monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL3
 
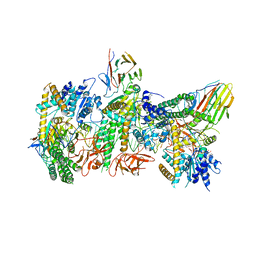 | | Cryo-EM structure of RIG-I:dsRNA filament in complex with RIPLET PrySpry domain (trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7LIU
 
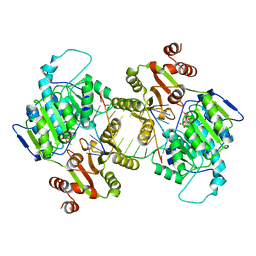 | | DDX3X bound to ATP analog and remodeled RNA:DNA hybrid | | Descriptor: | 5'-R(*GP*GP*GP*CP*GP*GP*G)-D(P*CP*CP*CP*GP*CP*CP*C)-3', ATP-dependent RNA helicase DDX3X, MAGNESIUM ION, ... | | Authors: | Enemark, E.J, Yu, S. | | Deposit date: | 2021-01-27 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | DDX3X bound to ATP analog and remodeled RNA:DNA hybrid
To Be Published
|
|
7LUV
 
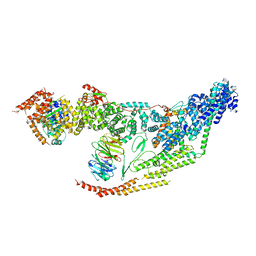 | | Cryo-EM structure of the yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2021-02-23 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast TREX complex and coordination with the SR-like protein Gbp2.
Elife, 10, 2021
|
|
7MQA
 
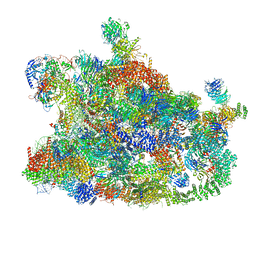 | | Cryo-EM structure of the human SSU processome, state post-A1 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Vanden Broeck, A, Singh, S, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
7MQJ
 
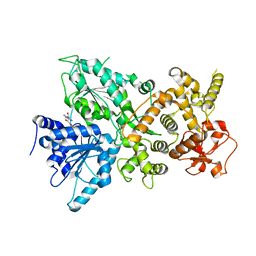 | | Dhr1 Helicase Core | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DHR1 | | Authors: | Miller, L, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
7NAC
 
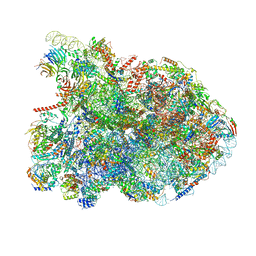 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Composite model | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NAD
 
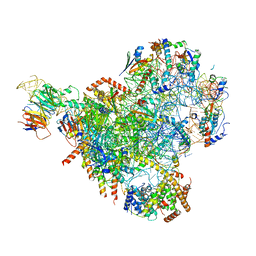 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Spb4 local refinement model | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L17-A, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OHP
 
 | |
7OHR
 
 | | Nog1-TAP associated immature ribosomal particle population E from S. cerevisiae | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Milkereit, P, Poell, G. | | Deposit date: | 2021-05-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.72 Å) | | Cite: | Analysis of subunit folding contribution of three yeast large ribosomal subunit proteins required for stabilisation and processing of intermediate nuclear rRNA precursors.
Plos One, 16, 2021
|
|
7OHS
 
 | |
7OHV
 
 | |
7OHW
 
 | |
7OHX
 
 | |
7OI6
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 1 | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-13 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
7OQB
 
 | | The U2 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | ACT1 pre-mRNA (delta-BS-A), Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing ATP-dependent RNA helicase PRP5, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQE
 
 | | Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A), Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
