2KFB
 
 | | The structure of the cataract causing P23T mutant of human gamma-D crystallin | | Descriptor: | Gamma-crystallin D | | Authors: | Jung, J, Byeon, I.L, Wang, Y, King, J, Gronenborn, A.M. | | Deposit date: | 2009-02-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the cataract-causing P23T mutant of human gammaD-crystallin exhibits distinctive local conformational and dynamic changes.
Biochemistry, 48, 2009
|
|
2KSG
 
 | |
2ORY
 
 | | Crystal structure of M37 lipase | | Descriptor: | Lipase | | Authors: | Jung, S.K, Jeong, D.G, Kim, S.J. | | Deposit date: | 2007-02-05 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cold adaptation of psychrophilic M37 lipase from Photobacterium lipolyticum.
Proteins, 71, 2008
|
|
1DEP
 
 | | MEMBRANE PROTEIN, NMR, 1 STRUCTURE | | Descriptor: | T345-359 | | Authors: | Jung, H, Schnackerz, K.D. | | Deposit date: | 1995-08-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR and circular dichroism studies of synthetic peptides derived from the third intracellular loop of the beta-adrenoceptor.
FEBS Lett., 358, 1995
|
|
2LN8
 
 | | The solution structure of theromacin | | Descriptor: | Theromacin | | Authors: | Jung, S, Soennichsen, F.D, Hung, C.-W, Tholey, A, Boidin-Wichlacz, C, Hausgen, W, Gelhaus, C, Desel, C, Podschun, R, Watzig, V, Tasiemski, A, Leippe, M, Groetzinger, J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Macin family of antimicrobial proteins combines antimicrobial and nerve repair activities.
J.Biol.Chem., 287, 2012
|
|
2LLR
 
 | |
384D
 
 | |
329D
 
 | |
382D
 
 | |
383D
 
 | |
3LPS
 
 | | Crystal structure of parE | | Descriptor: | NOVOBIOCIN, Topoisomerase IV subunit B | | Authors: | Jung, H.Y, Heo, Y.-S. | | Deposit date: | 2010-02-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of parE
To be Published
|
|
3L18
 
 | |
3HB0
 
 | | Structure of edeya2 complexed with bef3 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Eyes absent homolog 2 (Drosophila), MAGNESIUM ION | | Authors: | Jung, S.K, Jeong, D.G, Ryu, S.E, Kim, S.J. | | Deposit date: | 2009-05-03 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor
Faseb J., 24, 2010
|
|
3HB1
 
 | | Crystal structure of ed-eya2 complexed with Alf3 | | Descriptor: | ALUMINUM FLUORIDE, Eyes absent homolog 2 (Drosophila), MAGNESIUM ION | | Authors: | Jung, S.K, Jeong, D.G, Ryu, S.E, Kim, S.J. | | Deposit date: | 2009-05-03 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor
Faseb J., 24, 2010
|
|
5OQT
 
 | |
4D4G
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | APNAA1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D57
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, APNA A1, ARGININE, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4H
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | APNAA1, GLYCEROL | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4BPS
 
 | | Crystal structure of Chorismatase at 1.08 Angstrom resolution. | | Descriptor: | 3-(2-CARBOXYETHYL)BENZOIC ACID, FKBO | | Authors: | Juneja, P, Hubrich, F, Diederichs, K, Welte, W, Andexer, J.N. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-18 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | Mechanistic Implications for the Chorismatase Fkbo Based on the Crystal Structure.
J.Mol.Biol., 426, 2014
|
|
4D4I
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | APNAA1, ARGININE, GLYCEROL, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D56
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, APNAA1, GLYCEROL, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3R6S
 
 | |
3U55
 
 | | Crystal structure (Type-2) of SAICAR synthetase from Pyrococcus horikoshii OT3 | | Descriptor: | ACETATE ION, Phosphoribosylaminoimidazole-succinocarboxamide synthase, SULFATE ION | | Authors: | Manjunath, K, Kanaujia, S.P, Kanagaraj, S, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2011-10-11 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of SAICAR synthetase from Pyrococcus horikoshii OT3: insights into thermal stability
Int.J.Biol.Macromol., 53, 2013
|
|
3U54
 
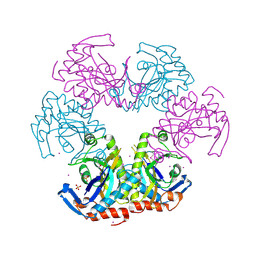 | | Crystal structure (Type-1) of SAICAR synthetase from Pyrococcus horikoshii OT3 | | Descriptor: | 1,4-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Manjunath, K, Kanaujia, S.P, Kanagaraj, S, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2011-10-11 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of SAICAR synthetase from Pyrococcus horikoshii OT3: insights into thermal stability
Int.J.Biol.Macromol., 53, 2013
|
|
7L69
 
 | |
