8OMQ
 
 | | DNA-unbound MutSbeta-ATP complex (straight clamp form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | DNA-unbound MutSbeta-ATP complex (straight clamp form)
To Be Published
|
|
8OMR
 
 | | Human tRNA guanine transglycosylase (TGT) bound to tRNAAsp | | Descriptor: | 9-DEAZAGUANINE, Queuine tRNA-ribosyltransferase accessory subunit 2, Queuine tRNA-ribosyltransferase catalytic subunit 1, ... | | Authors: | Sievers, K, Neumann, P, Susac, L, Trowitzsch, S, Tampe, R, Ficner, R. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insights into tRNA recognition by human tRNA guanine transglycosylase.
Structure, 32, 2024
|
|
4R33
 
 | | X-ray structure of the tryptophan lyase NosL with Tryptophan and S-adenosyl-L-homocysteine bound | | Descriptor: | CHLORIDE ION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nicolet, Y, Zeppieri, L, Amara, P, Fontecilla-Camps, J.-C. | | Deposit date: | 2014-08-14 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Tryptophan Lyase (NosL): Evidence for Radical Formation at the Amino Group of Tryptophan.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4R34
 
 | | X-ray structure of the tryptophan lyase NosL with Tryptophan, 5'-deoxyadenosine and methionine bound | | Descriptor: | 5'-DEOXYADENOSINE, BROMIDE ION, GLYCEROL, ... | | Authors: | Nicolet, Y, Zeppieri, L, Amara, P, Fontecilla-Camps, J.-C. | | Deposit date: | 2014-08-14 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Tryptophan Lyase (NosL): Evidence for Radical Formation at the Amino Group of Tryptophan.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5D1L
 
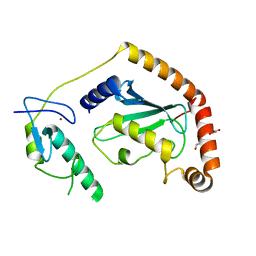 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 (Y165A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, OXALATE ION, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
5D1K
 
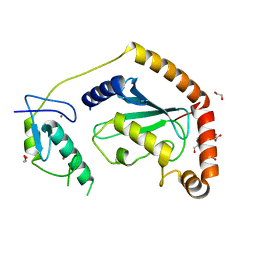 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
2XX9
 
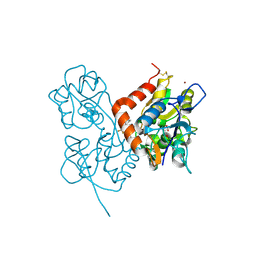 | | Crystal structure of 1-((2-fluoro-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XX7
 
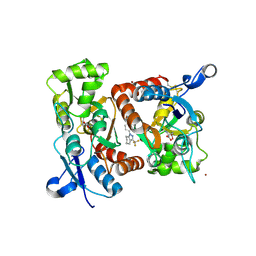 | | Crystal structure of 1-(4-(1-pyrrolidinylcarbonyl)phenyl)-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | 1-[4-(1-PYRROLIDINYLCARBONYL)PHENYL]-3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XXH
 
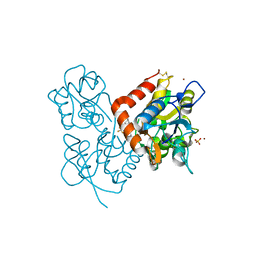 | | Crystal structure of 1-(4-(2-oxo-2-(1-pyrrolidinyl)ethyl)phenyl)-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.5A resolution. | | Descriptor: | 1-{4-[2-OXO-2-(1-PYRROLIDINYL)ETHYL]PHENYL}-3-( TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XX8
 
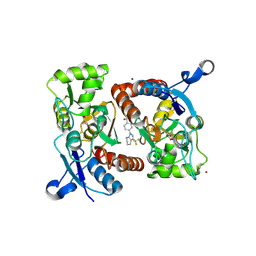 | | Crystal structure of N,N-dimethyl-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)benzamide in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-27 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
5CXX
 
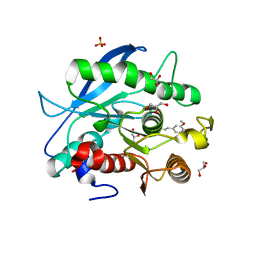 | | Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Gruninger, R.J, Abbott, D.W. | | Deposit date: | 2015-07-29 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Contributions of a unique beta-clamp to substrate recognition illuminates the molecular basis of exolysis in ferulic acid esterases.
Biochem.J., 473, 2016
|
|
4UPV
 
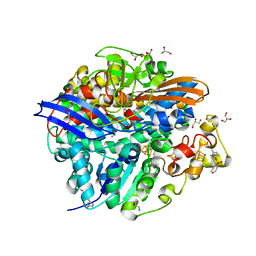 | | Low X-ray dose structure of a Ni-A Ni-Sox mixture of the D. fructosovorans NiFe-hydrogenase L122A mutant | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-18 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
5D1M
 
 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 (P199A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
5CXU
 
 | |
4UPE
 
 | | Structure of the unready Ni-A state of the S499C mutant of D. fructosovorans NiFe-hydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
2XXI
 
 | | Crystal structure of 1-((4-(3-(trifluoromethyl)-6,7-dihydropyrano(4,3- c(pyrazol-1(4H)-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.6A resolution. | | Descriptor: | 1-({4-[3-(TRIFLUOROMETHYL)-6,7-DIHYDROPYRANO[4,3-C]PYRAZOL-1(4H)-YL]PHENYL}METHYL)-2-PYRROLIDINONE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
7Y85
 
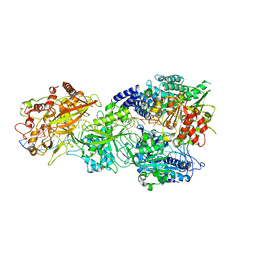 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease bound to self RNA target | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y81
 
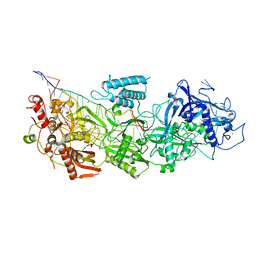 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA complex bound to non-self RNA target | | Descriptor: | MAGNESIUM ION, Non-self RNA target, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y82
 
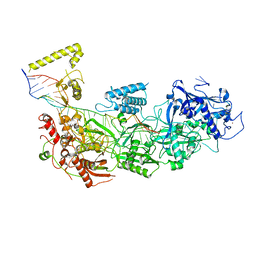 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA complex bound to self RNA target | | Descriptor: | MAGNESIUM ION, RAMP superfamily protein, Self RNA target, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y80
 
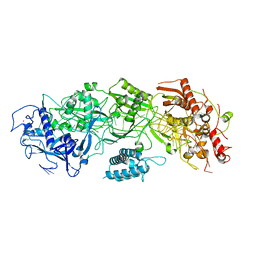 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA binary complex | | Descriptor: | MAGNESIUM ION, RAMP superfamily protein, ZINC ION, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y83
 
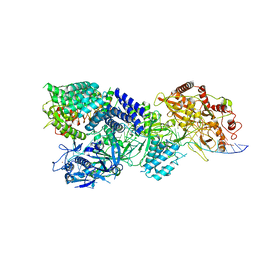 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease bound to non-self RNA target | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y84
 
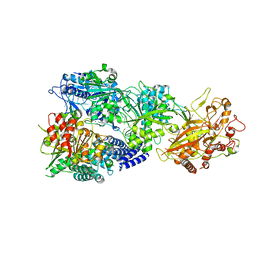 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
5B42
 
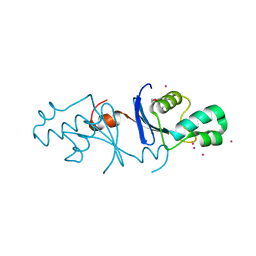 | | Crystal structure of the C-terminal endonuclease domain of Aquifex aeolicus MutL. | | Descriptor: | CADMIUM ION, DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL | | Authors: | Fukui, K, Baba, S, Kumasaka, T, Yano, T. | | Deposit date: | 2016-03-30 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Features and Functional Dependency on beta-Clamp Define Distinct Subfamilies of Bacterial Mismatch Repair Endonuclease MutL
J.Biol.Chem., 291, 2016
|
|
3AQF
 
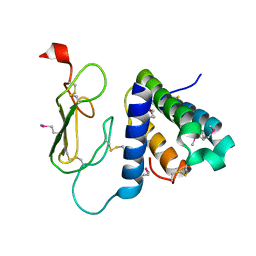 | | Crystal structure of the human CRLR/RAMP2 extracellular complex | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Receptor activity-modifying protein 2 | | Authors: | Kusano, S, Kukimono-Niino, M, Shirouzu, M, Shindo, T, Yokoyama, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding
Protein Sci., 21, 2012
|
|
4XP9
 
 | | X-ray structure of Drosophila dopamine transporter bound to psychostimulant D-amphetamine | | Descriptor: | (2S)-1-phenylpropan-2-amine, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
