3C9C
 
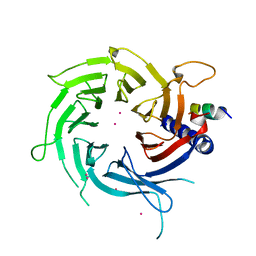 | | Structural Basis of Histone H4 Recognition by p55 | | 分子名称: | CADMIUM ION, Chromatin assembly factor 1 p55 subunit, Histone H4, ... | | 著者 | Song, J.J, Garlick, J.D, Kingston, R.E. | | 登録日 | 2008-02-15 | | 公開日 | 2008-05-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis of histone H4 recognition by p55.
Genes Dev., 22, 2008
|
|
3CFS
 
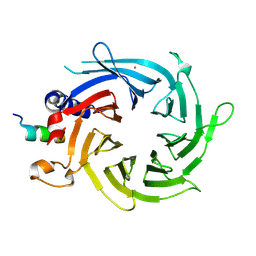 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | 分子名称: | ARSENIC, Histone H4, Histone-binding protein RBBP7 | | 著者 | Murzina, N.V, Pei, X.-Y, Pratap, J.V, Sparkes, M, Vicente-Garcia, J, Ben-Shahar, T.R, Verreault, A, Luisi, B.F, Laue, E.D. | | 登録日 | 2008-03-04 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
3CFV
 
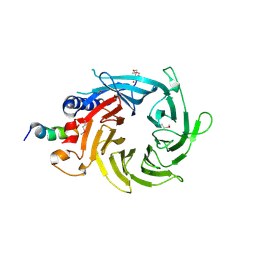 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | 分子名称: | ARSENIC, Histone H4 peptide, Histone-binding protein RBBP7 | | 著者 | Pei, X.-Y, Murzina, N.V, Zhang, W, McLaughlin, S, Verreault, A, Luisi, B.F, Laue, E.D. | | 登録日 | 2008-03-04 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
3DM0
 
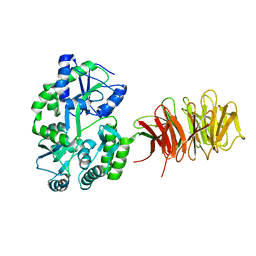 | | Maltose Binding Protein fusion with RACK1 from A. thaliana | | 分子名称: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein fused with RACK1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Ullah, H, Scappini, E.L, Moon, A.F, Williams, L.V, Armstrong, D.L, Pedersen, L.C. | | 登録日 | 2008-06-30 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana.
Protein Sci., 17, 2008
|
|
3EI2
 
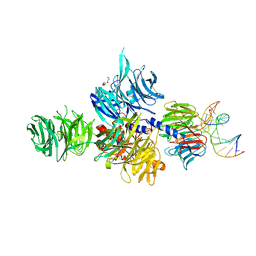 | |
3EI1
 
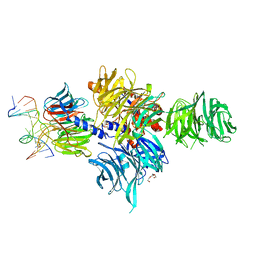 | |
3EI4
 
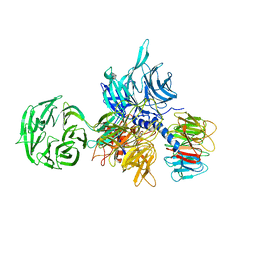 | |
3EI3
 
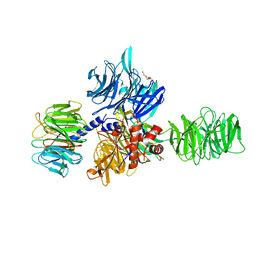 | | Structure of the hsDDB1-drDDB2 complex | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, TETRAETHYLENE GLYCOL | | 著者 | Scrima, A, Thoma, N.H. | | 登録日 | 2008-09-15 | | 公開日 | 2009-01-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3F3P
 
 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P21212 | | 分子名称: | Nucleoporin NUP85, Nucleoporin SEH1 | | 著者 | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | 登録日 | 2008-10-31 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
3EWE
 
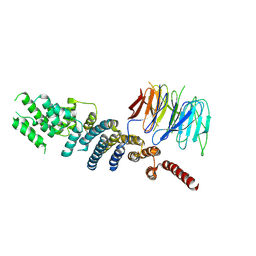 | | Crystal Structure of the Nup85/Seh1 Complex | | 分子名称: | Nucleoporin NUP85, Nucleoporin SEH1 | | 著者 | Brohawn, S.G, Leksa, N.C, Rajashankar, K.R, Schwartz, T.U. | | 登録日 | 2008-10-14 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural evidence for common ancestry of the nuclear pore complex and vesicle coats.
Science, 322, 2008
|
|
3F3F
 
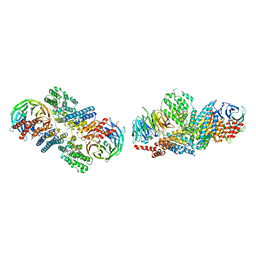 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P21 | | 分子名称: | Nucleoporin NUP85, Nucleoporin SEH1 | | 著者 | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | 登録日 | 2008-10-30 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
3F3G
 
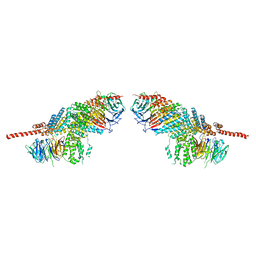 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P212121 | | 分子名称: | Nucleoporin NUP85, Nucleoporin SEH1 | | 著者 | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | 登録日 | 2008-10-30 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.75 Å) | | 主引用文献 | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
4R7A
 
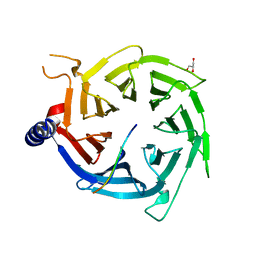 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | 分子名称: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | 著者 | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | 登録日 | 2014-08-27 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
4U7A
 
 | |
4UI9
 
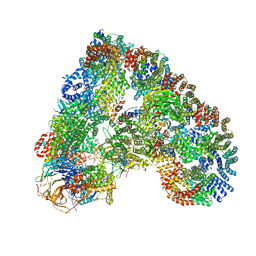 | | Atomic structure of the human Anaphase-Promoting Complex | | 分子名称: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | 著者 | Chang, L, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2015-03-27 | | 公開日 | 2015-06-17 | | 最終更新日 | 2019-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Nature, 522, 2015
|
|
5BJS
 
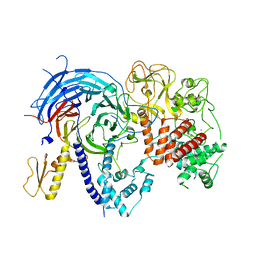 | | Apo ctPRC2 in an autoinhibited state | | 分子名称: | Histone-lysine N-methyltransferase EZH2, Polycomb protein SUZ12, Polycomb Protein EED, ... | | 著者 | Bratkowski, M.A, Liu, X. | | 登録日 | 2016-10-22 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.189 Å) | | 主引用文献 | Polycomb repressive complex 2 in an autoinhibited state.
J. Biol. Chem., 292, 2017
|
|
5C9Z
 
 | |
5CVL
 
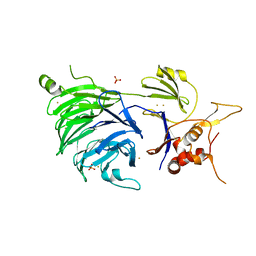 | | WDR48 (UAF-1), residues 2-580 | | 分子名称: | GOLD ION, PHOSPHATE ION, WD repeat-containing protein 48 | | 著者 | HARRIS, S.F, YIN, J. | | 登録日 | 2015-07-27 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
5CXB
 
 | |
5CXC
 
 | |
5CYK
 
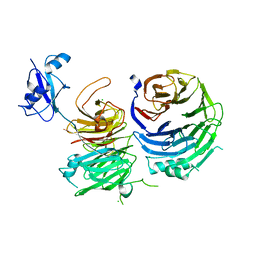 | | Structure of Ytm1 bound to the C-terminal domain of Erb1-R486E | | 分子名称: | CHLORIDE ION, Ribosome biogenesis protein ERB1, Ribosome biogenesis protein YTM1 | | 著者 | Wegrecki, M, Bravo, J. | | 登録日 | 2015-07-30 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two beta-propellers during the assembly of large ribosomal subunits in eukaryotes.
Nucleic Acids Res., 43, 2015
|
|
5EM2
 
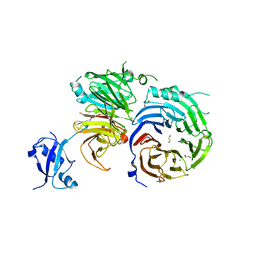 | | Crystal structure of the Erb1-Ytm1 complex | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ribosome biogenesis protein ERB1, ... | | 著者 | Ahmed, Y.L, Sinning, I. | | 登録日 | 2015-11-05 | | 公開日 | 2015-12-23 | | 最終更新日 | 2016-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface.
Nucleic Acids Res., 44, 2016
|
|
5EMM
 
 | |
5EML
 
 | |
5EMK
 
 | |
