1CSB
 
 | |
1HAO
 
 | | COMPLEX OF HUMAN ALPHA-THROMBIN WITH A 15MER OLIGONUCLEOTIDE GGTTGGTGTGGTTGG (BASED ON NMR MODEL OF DNA) | | Descriptor: | ALPHA-THROMBIN heavy chain, ALPHA-THROMBIN light chain, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Tulinsky, A, Padmanabhan, K. | | Deposit date: | 1995-10-03 | | Release date: | 1996-04-03 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An ambiguous structure of a DNA 15-mer thrombin complex.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1LAT
 
 | | GLUCOCORTICOID RECEPTOR MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*CP*CP*AP*GP*AP*AP*CP*AP*TP*GP*TP*TP*CP*TP*G P*GP*A)-3'), GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Gewirth, D.T, Sigler, P.B. | | Deposit date: | 1995-12-18 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The basis for half-site specificity explored through a non-cognate steroid receptor-DNA complex.
Nat.Struct.Biol., 2, 1995
|
|
1DIK
 
 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C.H. | | Deposit date: | 1995-12-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
241D
 
 | | EXTENSION OF THE FOUR-STRANDED INTERCALATED CYTOSINE MOTIF BY ADENINE.ADENINE BASE PAIRING IN THE CRYSTAL STRUCTURE OF D(CCCAAT) | | Descriptor: | DNA (5'-D(*CP*CP*CP*AP*AP*T)-3') | | Authors: | Berger, I, Kang, C, Fredian, A, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Extension of the four-stranded intercalated cytosine motif by adenine.adenine base pairing in the crystal structure of d(CCCAAT).
Nat.Struct.Biol., 2, 1995
|
|
1DVR
 
 | |
1AGI
 
 | |
1MMP
 
 | | MATRILYSIN COMPLEXED WITH CARBOXYLATE INHIBITOR | | Descriptor: | 5-METHYL-3-(9-OXO-1,8-DIAZA-TRICYCLO[10.6.1.013,18]NONADECA-12(19),13,15,17-TETRAEN-10-YLCARBAMOYL)-HEXANOIC ACID, CALCIUM ION, GELATINASE A, ... | | Authors: | Browner, M.F, Smith, W.W, Castelhano, A.L. | | Deposit date: | 1995-03-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Matrilysin-inhibitor complexes: common themes among metalloproteases.
Biochemistry, 34, 1995
|
|
1IRN
 
 | | RUBREDOXIN (ZN-SUBSTITUTED) AT 1.2 ANGSTROMS RESOLUTION | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Moulis, J.M, Meyer, J. | | Deposit date: | 1995-12-13 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Zinc- and iron-rubredoxins from Clostridium pasteurianum at atomic resolution: a high-precision model of a ZnS4 coordination unit in a protein.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1SLG
 
 | | STREPTAVIDIN, PH 5.6, BOUND TO PEPTIDE FCHPQNT | | Descriptor: | FCHPQNT, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1995-03-10 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Binding to protein targets of peptidic leads discovered by phage display: crystal structures of streptavidin-bound linear and cyclic peptide ligands containing the HPQ sequence
Biochemistry, 34, 1995
|
|
1SLE
 
 | | STREPTAVIDIN, PH 5.0, BOUND TO CYCLIC PEPTIDE AC-CHPQGPPC-NH2 | | Descriptor: | AC-CHPQGPPC-NH2, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1995-03-10 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding to protein targets of peptidic leads discovered by phage display: crystal structures of streptavidin-bound linear and cyclic peptide ligands containing the HPQ sequence
Biochemistry, 34, 1995
|
|
1SLD
 
 | |
1INO
 
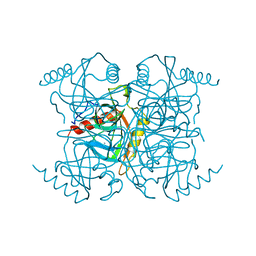 | |
1BI6
 
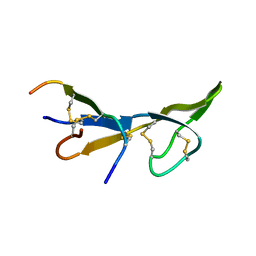 | | NMR STRUCTURE OF BROMELAIN INHIBITOR VI FROM PINEAPPLE STEM | | Descriptor: | BROMELAIN INHIBITOR VI | | Authors: | Hatano, K.-I. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of bromelain inhibitor IV from pineapple stem: structural similarity with Bowman-Birk trypsin/chymotrypsin inhibitor from soybean.
Biochemistry, 35, 1996
|
|
1CEN
 
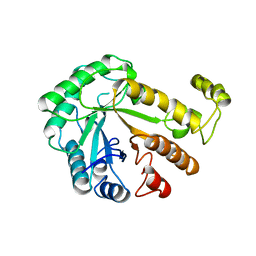 | |
2TCT
 
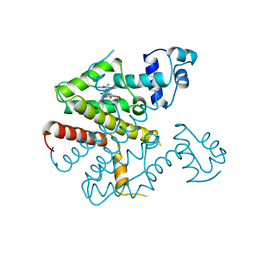 | |
1OEL
 
 | |
1CEO
 
 | |
1DMY
 
 | |
1APN
 
 | |
1SUT
 
 | |
180L
 
 | |
1RNQ
 
 | | RIBONUCLEASE A CRYSTALLIZED FROM 8M SODIUM FORMATE | | Descriptor: | FORMIC ACID, RIBONUCLEASE A | | Authors: | Fedorov, A.A, Josef-Mccarthy, D, Graf, I, Anguelova, D, Fedorov, E.V, Almo, S.C. | | Deposit date: | 1995-11-08 | | Release date: | 1996-04-03 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
1GTQ
 
 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | Descriptor: | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE, ZINC ION | | Authors: | Nar, H, Huber, R, Heizmann, C.W, Thoeny, B, Buergisser, D. | | Deposit date: | 1995-09-16 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of 6-pyruvoyl tetrahydropterin synthase, an enzyme involved in tetrahydrobiopterin biosynthesis.
EMBO J., 13, 1994
|
|
1VIH
 
 | | NMR STUDY OF VIGILIN, REPEAT 6, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
