7ZHH
 
 | | Complex structure of drosophila Unr CSD789 and a poly(A) RNA sequence | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, Upstream of N-ras, ... | | Authors: | Hollmann, N.M, Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res., 51, 2023
|
|
2ZI0
 
 | | Crystal structure of Tav2b/siRNA complex | | Descriptor: | Protein 2b, RNA (5'-D(P*AP*GP*AP*CP*AP*GP*CP*AP*UP*UP*AP*UP*GP*CP*UP*GP*UP*CP*UP*UP*U)-3') | | Authors: | Yuan, Y.A, Chen, H.-Y. | | Deposit date: | 2008-02-12 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b
Embo Rep., 9, 2008
|
|
1MNX
 
 | |
8DGA
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IV | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DFV
 
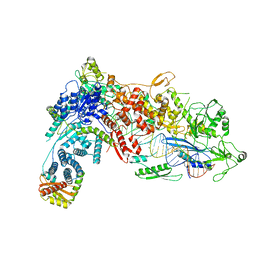 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIa | | Descriptor: | CALCIUM ION, Endoribonuclease Dcr-1, Loquacious, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-22 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DG5
 
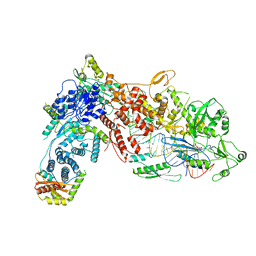 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIb | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
4MJ7
 
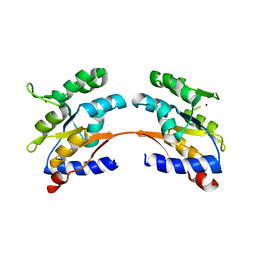 | |
4N0L
 
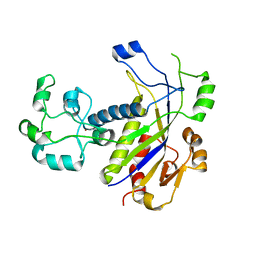 | | Methanopyrus kandleri Csm3 crystal structure | | Descriptor: | Predicted component of a thermophile-specific DNA repair system, contains a RAMP domain, ZINC ION | | Authors: | Hrle, A, Su, A.A, Ebert, J, Benda, C, Conti, E, Randau, L. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure and RNA-binding properties of the Type III-A CRISPR-associated protein Csm3.
Rna Biol., 10, 2013
|
|
2XLK
 
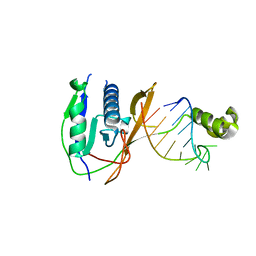 | | Crystal structure of the Csy4-crRNA complex, orthorhombic form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-21 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
7EIU
 
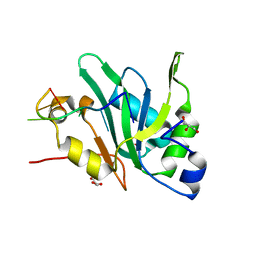 | | Crystal structure of Mei2 RRM3 in complex with 8mer meiRNA | | Descriptor: | GLYCEROL, Meiosis protein mei2, RNA (5'-R(P*UP*UP*CP*UP*GP*C)-3') | | Authors: | Shen, S.Y, Lv, M.Q. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structural insights reveal the specific recognition of meiRNA by the Mei2 protein.
J Mol Cell Biol, 14, 2022
|
|
2XLI
 
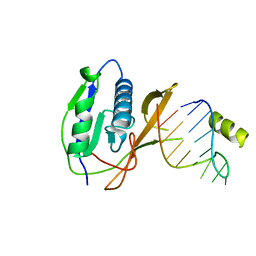 | | Crystal structure of the Csy4-crRNA complex, monoclinic form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
470D
 
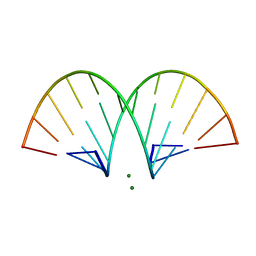 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
468D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
2XLJ
 
 | | Crystal structure of the Csy4-crRNA complex, hexagonal form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
469D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
471D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
3O3I
 
 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OH at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*U)-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-07-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4TXD
 
 | | Crystal structure of Thermofilum pendens Csc2 | | Descriptor: | Csc2, ZINC ION | | Authors: | Hrle, A, Conti, E. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Structural analyses of the CRISPR protein Csc2 reveal the RNA-binding interface of the type I-D Cas7 family.
Rna Biol., 11, 2014
|
|
310D
 
 | | Crystal structure of 2'-O-Me(CGCGCG)2: an RNA duplex at 1.3 A resolution. Hydration pattern of 2'-O-methylated RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(OMC)P*(OMG)P*(OMC)P*(OMG)P*(OMC)P*(OMG))-3') | | Authors: | Adamiak, D.A, Milecki, J, Adamiak, R.W, Dauter, Z, Rypniewski, W.R. | | Deposit date: | 1996-05-25 | | Release date: | 1997-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of 2'-O-Me(CGCGCG)2, an RNA duplex at 1.30 A resolution. Hydration pattern of 2'-O-methylated RNA.
Nucleic Acids Res., 25, 1997
|
|
4PDB
 
 | |
6D12
 
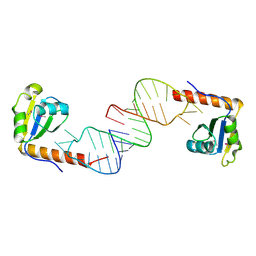 | | Crystal structure of C-terminal xRRM domain of human Larp7 bound to 7SK stem-loop 4 RNA | | Descriptor: | La-related protein 7, human 7SK RNA stem-loop 4 | | Authors: | Eichhorn, C.D, Cascio, D, Sawaya, M.R, Feigon, J. | | Deposit date: | 2018-04-11 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2L9W
 
 | |
3X1L
 
 | |
4AL5
 
 | |
