2KS6
 
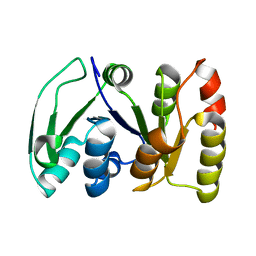 | |
1TCP
 
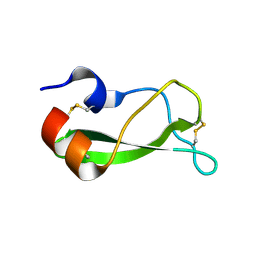 | |
1TBN
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
1TBO
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, 30 STRUCTURES | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
2L5Z
 
 | | NMR structure of the A730 loop of the Neurospora VS ribozyme | | Descriptor: | RNA (26-MER) | | Authors: | Desjardins, G, Bonneau, E, Girard, N, Boisbouvier, J, Legault, P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site.
Nucleic Acids Res., 39, 2011
|
|
2MIJ
 
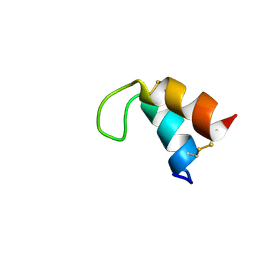 | | NMR structure of the S-linked glycopeptide sublancin 168 | | Descriptor: | SPBc2 prophage-derived bacteriocin sublancin-168, beta-D-glucopyranose | | Authors: | Garcia De Gonzalo, C.V, Zhu, L, Oman, T.J, van der Donk, W.A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-03-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the s-linked glycopeptide sublancin 168.
Acs Chem.Biol., 9, 2014
|
|
1S88
 
 | | NMR structure of a DNA duplex with two INA nucleotides inserted opposite each other, dCTCAACXCAAGCT:dAGCTTGXGTTGAG | | Descriptor: | 5'-D(*AP*GP*CP*TP*TP*GP*(2DM)P*GP*TP*TP*GP*AP*G)-3', 5'-D(*CP*TP*CP*AP*AP*CP*(2DM)P*CP*AP*AP*GP*CP*T)-3' | | Authors: | Nielsen, C.B, Petersen, M, Pedersen, E.B, Hansen, P.E, Christensen, U.B. | | Deposit date: | 2004-01-31 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a modified DNA oligonucleotide containing a new intercalating nucleic acid.
Bioconjug.Chem., 15, 2004
|
|
1TM6
 
 | |
2JVA
 
 | | NMR solution structure of peptidyl-tRNA hydrolase domain protein from Pseudomonas syringae pv. tomato. Northeast Structural Genomics Consortium target PsR211 | | Descriptor: | Peptidyl-tRNA hydrolase domain protein | | Authors: | Singarapu, K.K, Sukumaran, D, Parish, D, Eletsky, A, Zhang, Q, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the peptidyl-tRNA hydrolase domain from Pseudomonas syringae expands the structural coverage of the hydrolysis domains of class 1 peptide chain release factors.
Proteins, 71, 2008
|
|
2L9L
 
 | | NMR Structure of the Mouse MFG-E8 C2 Domain | | Descriptor: | Lactadherin | | Authors: | Ye, H, Yoon, H.S. | | Deposit date: | 2011-02-21 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of C2 domain of MFG-E8 and insights into its molecular recognition with phosphatidylserine
Biochim.Biophys.Acta, 1828, 2013
|
|
1TAP
 
 | | NMR SOLUTION STRUCTURE OF RECOMBINANT TICK ANTICOAGULANT PROTEIN (RTAP), A FACTOR XA INHIBITOR FROM THE TICK ORNITHODOROS MOUBATA | | Descriptor: | FACTOR XA INHIBITOR | | Authors: | Antuch, W, Guntert, P, Billeter, M, Wuthrich, K. | | Deposit date: | 1994-08-16 | | Release date: | 1994-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the recombinant tick anticoagulant protein (rTAP), a factor Xa inhibitor from the tick Ornithodoros moubata.
FEBS Lett., 352, 1994
|
|
1WAZ
 
 | |
2ITH
 
 | | NMR Structure of Haloferax volcanii DHFR | | Descriptor: | Dihydrofolate reductase | | Authors: | Binbuga, B. | | Deposit date: | 2006-10-19 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure in an extreme environment: NMR at high salt.
Protein Sci., 16, 2007
|
|
2KBL
 
 | |
2K48
 
 | | NMR Structure of the N-terminal Coiled Coil Domain of the Andes Hantavirus Nucleocapsid Protein | | Descriptor: | Nucleoprotein | | Authors: | Wang, Y, Boudreaux, D.M, Estrada, D.F, Egan, C.W, St Jeor, S.C, De Guzman, R.N. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the N-terminal Coiled Coil Domain of the Andes Hantavirus Nucleocapsid Protein.
J.Biol.Chem., 283, 2008
|
|
2L2F
 
 | |
2MIS
 
 | |
2I9H
 
 | |
2GG1
 
 | | NMR solution structure of domain III of the E-protein of tick-borne Langat flavivirus (includes RDC restraints) | | Descriptor: | Genome polyprotein | | Authors: | Mukherjee, M, Dutta, K, White, M.A, Cowburn, D, Fox, R.O. | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-25 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition.
Protein Sci., 15, 2006
|
|
2D3J
 
 | |
2CHJ
 
 | | NMR structure of TGLGLT quadruplex | | Descriptor: | 5'-D(*TP*G LCGP*G LCGP*TP)-3' | | Authors: | Nielsen, J.T, Petersen, M. | | Deposit date: | 2006-03-15 | | Release date: | 2006-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of Lna (Locked Nucleic Acid) Modified Quadruplexes
Nucleic Acids Res., 34, 2006
|
|
2CHK
 
 | | NMR structure of TLLLLT quadruplex | | Descriptor: | 5'-D(*T LCG LCG LCG LCGP*TP)-3' | | Authors: | Nielsen, J.T, Petersen, M. | | Deposit date: | 2006-03-15 | | Release date: | 2006-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of Lna (Locked Nucleic Acid) Modified Quadruplexes
Nucleic Acids Res., 34, 2006
|
|
2CKN
 
 | |
2HEM
 
 | | NMR structure and Mg2+ binding of an RNA segment that underlies the L7/L12 stalk in the E.coli 50S ribosomal subunit. | | Descriptor: | 5'-R(P*GP*GP*GP*AP*AP*GP*GP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*CP*GP*GP*CP*CP*C)-3' | | Authors: | Zhao, Q, Nagaswamy, U, Lee, H, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2006-06-21 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and Mg2+ binding of an RNA segment that underlies the L7/L12 stalk in the E.coli 50S ribosomal subunit
Nucleic Acids Res., 33, 2005
|
|
2IGZ
 
 | |
