7G5M
 
 | | Crystal Structure of rat Autotaxin in complex with rac-(E)-1-[rac-(3aR,8aS)-2-(1,4,6,7-tetrahydrotriazolo[4,5-c]pyridine-5-carbonyl)-1,3,3a,4,5,7,8,8a-octahydropyrrolo[3,4-d]azepin-6-yl]-3-[4-(trifluoromethoxy)phenyl]prop-2-en-1-one, i.e. SMILES C1C[C@@H]2[C@H](CCN1C(=O)/C=C/c1ccc(cc1)OC(F)(F)F)CN(C2)C(=O)N1CCC2=C(C1)N=NN2 with IC50=0.0280543 microM | | Descriptor: | (2E)-1-[(3aS,8aS)-2-(1,4,6,7-tetrahydro-5H-[1,2,3]triazolo[4,5-c]pyridine-5-carbonyl)octahydropyrrolo[3,4-d]azepin-6(1H)-yl]-3-[4-(trifluoromethoxy)phenyl]prop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3PR0
 
 | | Crystal Structure of a Covalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | 7-phenyl-1-[5-(pyridin-2-yl)-1,3,4-oxadiazol-2-yl]heptane-1,1-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mileni, M, Han, G.W, Boger, D.L, Stevens, R.C. | | Deposit date: | 2010-11-29 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fluoride-mediated capture of a noncovalent bound state of a reversible covalent enzyme inhibitor: X-ray crystallographic analysis of an exceptionally potent alpha-ketoheterocycle inhibitor of fatty acid amide hydrolase.
J.Am.Chem.Soc., 133, 2011
|
|
3VD9
 
 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
4I3N
 
 | |
3VDC
 
 | | E. coli (lacZ) beta-galactosidase (N460T) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
1IEV
 
 | | CRYSTAL STRUCTURE OF BARLEY BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1 IN COMPLEX WITH CYCLOHEXITOL | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hrmova, M, DeGori, R, Fincher, G.B, Varghese, J.N. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
1J0J
 
 | |
3QWJ
 
 | | CDK2 in complex with inhibitor KVR-1-142 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-aminopyrimidin-5-yl)methyl]amino}-4-chloro-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QX4
 
 | | CDK2 in complex with inhibitor KVR-1-78 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-01 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QX2
 
 | | CDK2 in complex with inhibitor KVR-1-190 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-aminopyrimidin-5-yl)methyl]amino}-4-[(2-hydroxyethyl)amino]-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-01 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QZI
 
 | | CDK2 in complex with inhibitor KVR-1-126 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-5-nitro-2-[(pyrimidin-5-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-06 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
1UKC
 
 | | Crystal Structure of Aspergillus niger EstA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bourne, Y, Hasper, A.A, Chahinian, H, Juin, M, De Graaff, L.H, Marchot, P. | | Deposit date: | 2003-08-19 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aspergillus niger protein EstA defines a new class of fungal esterases within the alpha/beta hydrolase fold superfamily of proteins
STRUCTURE, 12, 2004
|
|
3QXO
 
 | | CDK2 in complex with inhibitor KVR-1-84 | | Descriptor: | 1,2-ETHANEDIOL, 5-nitro-2-[(4-sulfamoylbenzyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-02 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3VHV
 
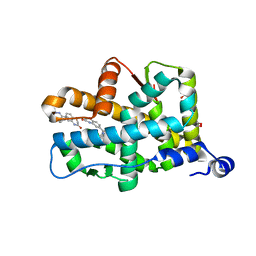 | | Mineralocorticoid receptor ligand-binding domain with non-steroidal antagonist | | Descriptor: | 1,2-ETHANEDIOL, 6-[(1E)-2-phenyl-N-(3-sulfanyl-4H-1,2,4-triazol-4-yl)ethanimidoyl]-2H-1,4-benzoxazin-3(4H)-one, 6-[(7S)-7-phenyl-7H-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazin-6-yl]-2H-1,4-benzoxazin-3(4H)-one, ... | | Authors: | Sogabe, S, Habuka, N. | | Deposit date: | 2011-09-07 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of Benzoxazin-3-one Derivatives as Novel, Potent, and Selective Nonsteroidal Mineralocorticoid Receptor Antagonists
J.Med.Chem., 54, 2011
|
|
2OVG
 
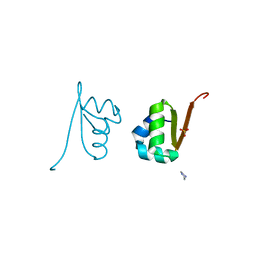 | | Lambda Cro Q27P/A29S/K32Q triple mutant at 1.35 A in space group P3221 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Phage lambda Cro, SULFATE ION | | Authors: | Hall, B.M, Heroux, A, Roberts, S.A, Cordes, M.H. | | Deposit date: | 2007-02-13 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Two structures of a lambda Cro variant highlight dimer flexibility but disfavor major dimer distortions upon specific binding of cognate DNA.
J.Mol.Biol., 375, 2008
|
|
2AC1
 
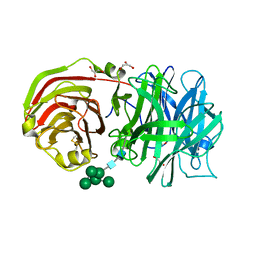 | | Crystal structure of a cell-wall invertase from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Verhaest, M, Le Roy, K, De Ranter, C, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2005-07-18 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray diffraction structure of a cell-wall invertase from Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2ACH
 
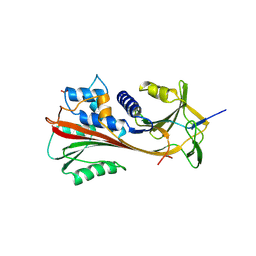 | | CRYSTAL STRUCTURE OF CLEAVED HUMAN ALPHA1-ANTICHYMOTRYPSIN AT 2.7 ANGSTROMS RESOLUTION AND ITS COMPARISON WITH OTHER SERPINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA 1-ANTICHYMOTRYPSIN, PHOSPHATE ION, ... | | Authors: | Baumann, U, Huber, R, Bode, W, Grosse, D, Lesjak, M, Laurell, C.B. | | Deposit date: | 1993-04-26 | | Release date: | 1993-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cleaved human alpha 1-antichymotrypsin at 2.7 A resolution and its comparison with other serpins.
J.Mol.Biol., 218, 1991
|
|
3QZH
 
 | | CDK2 in complex with inhibitor KVR-1-124 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-06 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R6X
 
 | | CDK2 in complex with inhibitor KVR-1-158 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-hydroxypropyl)amino]-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-22 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R7U
 
 | | CDK2 in complex with inhibitor KVR-1-75 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(aminomethyl)benzyl]amino}-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R7V
 
 | | CDK2 in complex with inhibitor KVR-1-9 | | Descriptor: | 1,2-ETHANEDIOL, 5-nitro-2-[(pyridin-2-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R71
 
 | | CDK2 in complex with inhibitor KVR-1-162 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-methoxypropyl)amino]-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-22 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3RPO
 
 | | CDK2 in complex with inhibitor KVR-1-156 | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-carbamoyl-2-nitro-5-[(pyridin-3-ylmethyl)amino]phenyl}amino)butanoic acid, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-27 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
2DV9
 
 | | Crystal structure of peanut lectin GAL-BETA-1,3-GAL complex | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
3RM7
 
 | | CDK2 in complex with inhibitor KVR-1-91 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4-hydroxybenzyl)amino]-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
