5S8C
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N00322e (space group C2) | | Descriptor: | (1-methylbenzotriazol-5-yl)methanol, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8L
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with E11289c (space group C2) | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8P
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with E07179c (space group P212121) | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-3,4-dihydroquinazolin-2(1H)-one, CALCIUM ION, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | XChem group deposition
To Be Published
|
|
2Z36
 
 | | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome P450 type compactin 3'',4''-hydroxylase, FE (III) ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Fujii, T, Arisawa, A, Tamura, T. | | Deposit date: | 2007-06-02 | | Release date: | 2007-08-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105)
Biochem.Biophys.Res.Commun., 361, 2007
|
|
6B5J
 
 | | TNNI3K complexed with a 4,6-diaminopyrimidine | | Descriptor: | N-methyl-3-[(6-{[4-(trifluoromethyl)phenyl]amino}pyrimidin-4-yl)amino]benzene-1-sulfonamide, Serine/threonine-protein kinase TNNI3K | | Authors: | Shewchuk, L.M, Philp, J. | | Deposit date: | 2017-09-29 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | 4,6-Diaminopyrimidines as Highly Preferred Troponin I-Interacting Kinase (TNNI3K) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5UR6
 
 | | PYR1 bound to the rationally designed agonist cyanabactin | | Descriptor: | Abscisic acid receptor PYR1, N-(4-cyano-3-cyclopropylphenyl)-1-(4-methylphenyl)methanesulfonamide, SULFATE ION | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2017-02-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | A Rationally Designed Agonist Defines Subfamily IIIA Abscisic Acid Receptors As Critical Targets for Manipulating Transpiration.
ACS Chem. Biol., 12, 2017
|
|
7TAE
 
 | |
5M9D
 
 | |
4H22
 
 | |
5J6T
 
 | |
5J6V
 
 | |
5J6W
 
 | |
8SLM
 
 | | Crystal structure of Deinococcus geothermalis PprI | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Zn dependent hydrolase fused to HTH domain, ... | | Authors: | Zhao, Y, Lu, H. | | Deposit date: | 2023-04-23 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The Deinococcus protease PprI senses DNA damage by directly interacting with single-stranded DNA.
Nat Commun, 15, 2024
|
|
8SLN
 
 | |
1FQ1
 
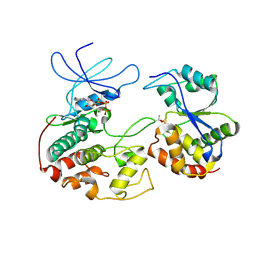 | | CRYSTAL STRUCTURE OF KINASE ASSOCIATED PHOSPHATASE (KAP) IN COMPLEX WITH PHOSPHO-CDK2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-DEPENDENT KINASE INHIBITOR 3, ... | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
1FEP
 
 | | FERRIC ENTEROBACTIN RECEPTOR | | Descriptor: | FERRIC ENTEROBACTIN RECEPTOR | | Authors: | Buchanan, S.K, Smith, B.S, Ventatramani, L, Xia, D, Esser, L, Palnitkar, M, Chakraborty, R, Van Der Helm, D, Deisenhofer, J. | | Deposit date: | 1998-11-24 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the outer membrane active transporter FepA from Escherichia coli.
Nat.Struct.Biol., 6, 1999
|
|
5ZUJ
 
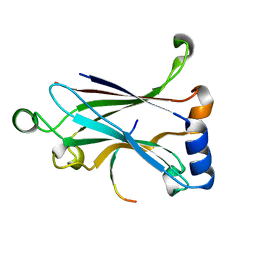 | | Binding and Enhanced Binding between Key Immunity Proteins TRAF6 and TIFA | | Descriptor: | TNF receptor-associated factor 6, peptide 170-184 from TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Huang, W.C, Maestre-Reyna, M, Hsiao, T.C, Tsai, M.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding and Enhanced Binding between Key Immunity Proteins TRAF6 and TIFA.
Chembiochem, 20, 2019
|
|
5LIL
 
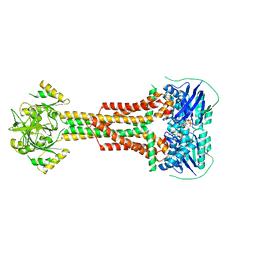 | | Structure of Aggregatibacter actinomycetemcomitans MacB bound to ATPyS (P21) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Macrolide export ATP-binding/permease protein MacB | | Authors: | Crow, A. | | Deposit date: | 2016-07-15 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LIR
 
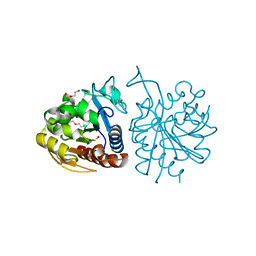 | |
8CIH
 
 | | Structure of FL CINP | | Descriptor: | Cyclin-dependent kinase 2-interacting protein, SODIUM ION | | Authors: | Foglizzo, M, Zeqiraj, E. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity.
Cell, 187, 2024
|
|
5LVF
 
 | | Solution structure of Rtt103 CTD-interacting domain bound to a Thr4 phosphorylated CTD peptide | | Descriptor: | PRO-SER-TYR-SER-PRO-PTH-SER-PRO-SER-TYR-SER-PRO-THR-SER-PRO-SER, Regulator of Ty1 transposition protein 103 | | Authors: | Jasnovidova, O, Kubicek, K, Krejcikova, M, Stefl, R. | | Deposit date: | 2016-09-14 | | Release date: | 2017-05-10 | | Last modified: | 2019-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural insight into recognition of phosphorylated threonine-4 of RNA polymerase II C-terminal domain by Rtt103p.
EMBO Rep., 18, 2017
|
|
7JUA
 
 | |
7JTJ
 
 | |
6BEK
 
 | |
5MDT
 
 | |
