4P0D
 
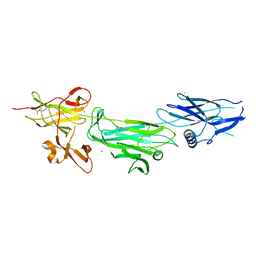 | | The T6 backbone pilin of serotype M6 Streptococcus pyogenes has a modular three-domain structure decorated with variable loops and extensions | | Descriptor: | CALCIUM ION, IODIDE ION, Trypsin-resistant surface T6 protein | | Authors: | Young, P.G, Moreland, N.J, Loh, J.M, Bell, A, Atatoa-Carr, P, Proft, T, Baker, E.N. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Conservation, Variability, and Immunogenicity of the T6 Backbone Pilin of Serotype M6 Streptococcus pyogenes.
Infect.Immun., 82, 2014
|
|
8SQX
 
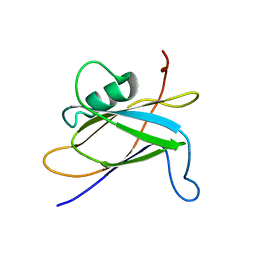 | |
4BHQ
 
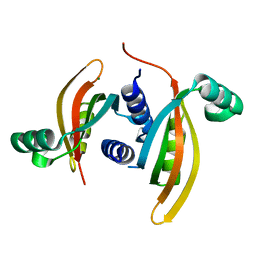 | | Structure of the periplasmic domain of the PilN type IV pilus biogenesis protein from Thermus thermophilus | | Descriptor: | COMPETENCE PROTEIN PILN, MAGNESIUM ION | | Authors: | Karuppiah, V, Collins, R.F, Gao, Y, Thistlethwaite, A, Derrick, J.P. | | Deposit date: | 2013-04-05 | | Release date: | 2013-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Assembly of an Inner Membrane Platform for Initiation of Type Iv Pilus Biogenesis
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2JMR
 
 | | NMR structure of the E. coli type 1 pilus subunit FimF | | Descriptor: | fimF | | Authors: | Gossert, A.D, Bettendorff, P, Puorger, C, Vetsch, M, Herrmann, T, Fiorito, F, Hiller, S, Glockshuber, R, Wuthrich, K. | | Deposit date: | 2006-11-29 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Escherichia coli type 1 pilus subunit FimF and its interactions with other pilus subunits.
J.Mol.Biol., 375, 2008
|
|
6FQA
 
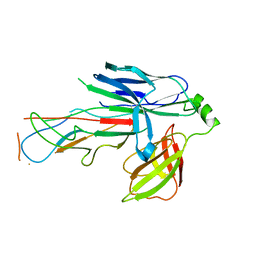 | | Crystal structure of the CsuC-CsuA/B chaperone-subunit preassembly complex of the archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B, CsuC | | Authors: | Parilova, O, Pakharukova, N.A, Malmi, H, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-02-13 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
4K8W
 
 | | An arm-swapped dimer of the S. pyogenes pilin specific assembly factor SipA | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, LepA | | Authors: | Young, P.G, Kang, H.J, Baker, E.N. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An arm-swapped dimer of the Streptococcus pyogenes pilin specific assembly factor SipA.
J.Struct.Biol., 183, 2013
|
|
4OGM
 
 | | MBP-fusion protein of PilA1 residues 26-159 | | Descriptor: | Maltose ABC transporter periplasmic protein, pilin protein chimera, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
1RG0
 
 | |
4PE2
 
 | | MBP PilA1 CD160 | | Descriptor: | MALONATE ION, Maltose ABC transporter periplasmic protein,Prepilin-type N-terminal cleavage/methylation domain protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-04-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
4D40
 
 | | High-Resolution Structure of a Type IV Pilin from Shewanella oneidensis | | Descriptor: | PILD PROCESSED PROTEIN, SODIUM ION, SULFATE ION | | Authors: | Gorgel, M, JensenUlstrup, J, Boeggild, A, Nissen, P, Boesen, T. | | Deposit date: | 2014-10-24 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.669 Å) | | Cite: | High-Resolution Structure of a Type Iv Pilin from the Metal- Reducing Bacterium Shewanella Oneidensis.
Bmc Struct.Biol., 15, 2015
|
|
2YCH
 
 | | PilM-PilN type IV pilus biogenesis complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COMPETENCE PROTEIN PILM, COMPETENCE PROTEIN PILN, ... | | Authors: | Karuppiah, V, Derrick, J.P. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-11 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Pilm-Piln Inner Membrane Type Iv Pilus Biogenesis Complex from Thermus Thermophilus.
J.Biol.Chem., 286, 2011
|
|
6FM5
 
 | | Crystal structure of self-complemented CsuA/B major subunit from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B,CsuA/B,CsuA/B | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
6FQ0
 
 | | Crystal structure of the CsuC-CsuA/B chaperone-subunit preassembly complex of the archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B, CsuC | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-02-12 | | Release date: | 2018-09-26 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
1QVE
 
 | |
8RJF
 
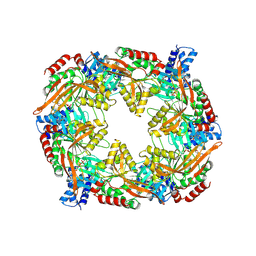 | | TadA/CpaF with ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hohl, M, Low, H. | | Deposit date: | 2023-12-21 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bidirectional pilus processing in the Tad pilus system motor CpaF.
Nat Commun, 15, 2024
|
|
8RKD
 
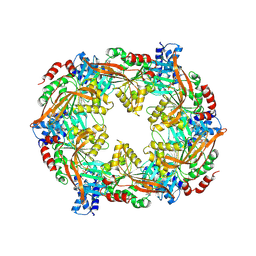 | | TadA/CpaF with AMPPNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hohl, M, Low, H. | | Deposit date: | 2023-12-24 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Bidirectional pilus processing in the Tad pilus system motor CpaF.
Nat Commun, 15, 2024
|
|
8RKL
 
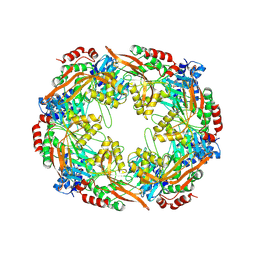 | | TadA/CpaF nucleotide free | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Pilus assembly ATPase CpaF | | Authors: | Hohl, M, Low, H. | | Deposit date: | 2023-12-26 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Bidirectional pilus processing in the Tad pilus system motor CpaF.
Nat Commun, 15, 2024
|
|
4C0Z
 
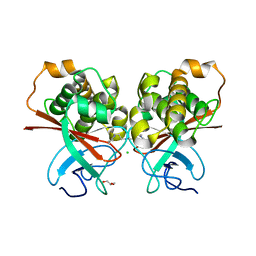 | | The N-terminal domain of the Streptococcus pyogenes pilus tip adhesin Cpa | | Descriptor: | ANCILLARY PROTEIN 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Linke-Winnebeck, C, Paterson, N, Baker, E.N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-11-20 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Model for the Covalent Adhesion of the Streptococcus Pyogenes Pilus Through a Thioester Bond.
J.Biol.Chem., 289, 2014
|
|
2PY0
 
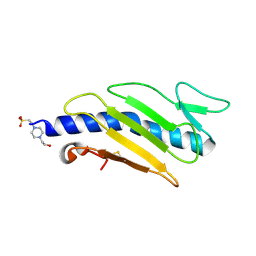 | | Crystal structure of Cs1 pilin chimera | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fimbrial protein | | Authors: | Kao, D.J, Churchill, M.E, Hodges, R.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Animal Protection and Structural Studies of a Consensus Sequence Vaccine Targeting the Receptor Binding Domain of the Type IV Pilus of Pseudomonas aeruginosa.
J.Mol.Biol., 374, 2007
|
|
2FI7
 
 | |
2Y1V
 
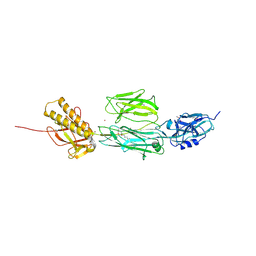 | | Full length structure of RrgB Pilus protein from Streptococcus pneumoniae | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, NICKEL (II) ION | | Authors: | El-Mortaji, L, Contreras-Martel, C, Manzano, C, Vernet, T, Dessen, A, DiGuilmi, A.M. | | Deposit date: | 2010-12-10 | | Release date: | 2011-11-09 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The Full-Length Streptococcus Pneumoniae Major Pilin Rrgb Crystallizes in a Fibre-Like Structure, which Presents the D1 Isopeptide Bond and Provides Details on the Mechanism of Pilus Polymerization.
Biochem.J., 441, 2012
|
|
6OLJ
 
 | |
6OJY
 
 | |
6OJX
 
 | |
6OKV
 
 | |
