5HBD
 
 | | Filamentous Assembly of Green Fluorescent Protein Supported by a C-terminal fusion of 18-residues, viewed in space group C2 | | Descriptor: | Green fluorescent protein | | Authors: | Sawaya, M.R, Hochschild, A, Heller, D.M, McPartland, L, Eisenberg, D.S. | | Deposit date: | 2015-12-31 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Green Fluorescent Protein Fusion that Self Assembles as Polar Filaments
to be published
|
|
7SFA
 
 | |
6QSM
 
 | |
5EB7
 
 | |
4TPS
 
 | | Sporulation Inhibitor of DNA Replication, SirA, in complex with Domain I of DnaA | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, Chromosomal replication initiator protein DnaA, ... | | Authors: | Jameson, K.H, Turkenburg, J.P, Fogg, M.J, Grahl, A, Wilkinson, A.J. | | Deposit date: | 2014-06-09 | | Release date: | 2014-07-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and interactions of the Bacillus subtilis sporulation inhibitor of DNA replication, SirA, with domain I of DnaA.
Mol.Microbiol., 93, 2014
|
|
2A46
 
 | |
5EJU
 
 | |
5EXU
 
 | |
3U0M
 
 | |
5EB6
 
 | |
5K9C
 
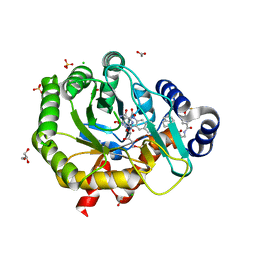 | | Crystal structure of human dihydroorotate dehydrogenase with ML390 | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lewis, T.A, Sykes, D.B, Law, J.M, Munoz, B, Scadden, D.T, Rustiguel, J.K, Nonato, M.C, Schreiber, S.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia.
ACS Med Chem Lett, 7, 2016
|
|
6QQA
 
 | |
4L13
 
 | |
5MSE
 
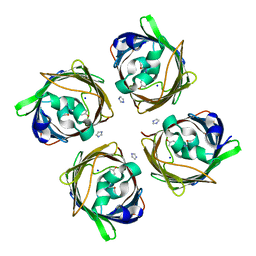 | | GFP nuclear transport receptor mimic 3B8 | | Descriptor: | Green fluorescent protein, IMIDAZOLE, SODIUM ION | | Authors: | Huyton, T, Gorlich, D. | | Deposit date: | 2017-01-04 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Surface Properties Determining Passage Rates of Proteins through Nuclear Pores.
Cell, 174, 2018
|
|
6LR7
 
 | |
3RWA
 
 | |
6GO9
 
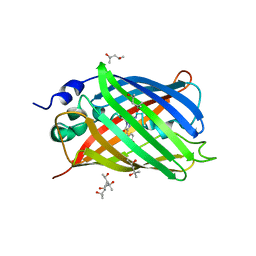 | | Structure of GFPmut2 crystallized at pH 6 and transferred to pH 7 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Green fluorescent protein | | Authors: | Lolli, G, Raboni, S, Pasqualetto, E, Campanini, B, Mozzarelli, A, Bettati, S, Battistutta, R. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.672 Å) | | Cite: | Insight into GFPmut2 pH Dependence by Single Crystal Microspectrophotometry and X-ray Crystallography.
J.Phys.Chem.B, 122, 2018
|
|
6QUJ
 
 | | GHK tagged GFP variant | | Descriptor: | COPPER (II) ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Huyton, T, Gorlich, D. | | Deposit date: | 2019-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1OXF
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
6B9C
 
 | | Superfolder Green Fluorescent Protein with 4-nitro-L-phenylalanine at the chromophore (position 66) | | Descriptor: | CARBON DIOXIDE, Green fluorescent protein | | Authors: | Phillips-Piro, C.M, Brewer, S.H, Olenginski, G.M, Piacentini, J. | | Deposit date: | 2017-10-10 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
3DQ1
 
 | |
2XVY
 
 | | Cobalt chelatase CbiK (periplasmic) from Desulvobrio vulgaris Hildenborough (co-crystallised with cobalt and SHC) | | Descriptor: | CHELATASE, PUTATIVE, COBALT (II) ION, ... | | Authors: | Romao, C.V, Lobo, S.A.L, Carrondo, M.A, Saraiva, L.M, Matias, P.M. | | Deposit date: | 2010-10-28 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Desulfovibrio vulgaris CbiK(P) cobaltochelatase: evolution of a haem binding protein orchestrated by the incorporation of two histidine residues.
Environ. Microbiol., 19, 2017
|
|
5K9D
 
 | | Crystal structure of human dihydroorotate dehydrogenase at 1.7 A resolution | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lewis, T.A, Sykes, D.B, Law, J.M, Munoz, B, Scadden, D.T, Rustiguel, J.K, Nonato, M.C, Schreiber, S.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia.
ACS Med Chem Lett, 7, 2016
|
|
4IR5
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-{1-[2-(hydroxymethyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone (GSK2847449A) | | Descriptor: | 1,2-ETHANEDIOL, 1-{1-[2-(hydroxymethyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
3DQ3
 
 | |
