2BL0
 
 | | Physarum polycephalum myosin II regulatory domain | | Descriptor: | CALCIUM ION, MAJOR PLASMODIAL MYOSIN HEAVY CHAIN, MYOSIN REGULATORY LIGHT CHAIN | | Authors: | Debreczeni, J.E, Farkas, L, Harmat, V, Nyitray, L. | | Deposit date: | 2005-02-23 | | Release date: | 2005-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Evidence for Non-Canonical Binding of Ca2+ to a Canonical EF-Hand of a Conventional Myosin.
J.Biol.Chem., 280, 2005
|
|
2BL1
 
 | | Crystal structure of a putative phosphinothricin Acetyltransferase (PA4866) from Pseudomonas aeruginosa PAC1 | | Descriptor: | AZIDE ION, GLYCEROL, PUTATIVE PHOSPHINOTHRICIN N-ACETYLTRANSFERASE PA4866, ... | | Authors: | Davies, A.M, Tata, R, Agha, R, Sutton, B.J, Brown, P.R. | | Deposit date: | 2005-02-24 | | Release date: | 2005-09-21 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Putative Phosphinothricin Acetyltransferase (Pa4866) from Pseudomonas Aeruginosa Pac1
Proteins: Struct., Funct., Bioinf., 61, 2005
|
|
2BL2
 
 | | The membrane rotor of the V-type ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, SODIUM ION, UNDECYL-MALTOSIDE, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2005-02-25 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Rotor of the Vacuolar-Type Na- ATPase from Enterococcus Hirae
Science, 308, 2005
|
|
2BL4
 
 | | Lactaldehyde:1,2-propanediol oxidoreductase of Escherichia coli | | Descriptor: | CHLORIDE ION, FE (II) ION, LACTALDEHYDE REDUCTASE, ... | | Authors: | Montella, C, Bellsolell, L, Perez-Luque, R, Badia, J, Baldoma, L, Coll, M, Aguilar, J. | | Deposit date: | 2005-03-01 | | Release date: | 2005-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of an Iron-Dependent Group III Dehydrogenase that Interconverts L-Lactaldehyde and L-1,2-Propanediol in Escherichia Coli.
J.Bacteriol., 187, 2005
|
|
2BL5
 
 | | Solution structure of the KH-QUA2 region of the Xenopus STAR-GSG Quaking protein. | | Descriptor: | MGC83862 PROTEIN | | Authors: | Maguire, M.L, Guler-Gane, G, Nietlispach, D, Raine, A.R.C, Zorn, A.M, Standart, N, Broadhurst, R.W. | | Deposit date: | 2005-03-01 | | Release date: | 2005-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Kh-Qua2 Region of the Xenopus Star/Gsg Quaking Protein
J.Mol.Biol., 348, 2005
|
|
2BL6
 
 | | Solution structure of the Zn complex of EIAV NCp11(22-58) peptide, including two CCHC Zn-binding motifs. | | Descriptor: | NUCLEOCAPSID PROTEIN P11, ZINC ION | | Authors: | Amodeo, P, Castiglione-Morelli, M.A, Ostuni, A, Bavoso, A. | | Deposit date: | 2005-03-02 | | Release date: | 2006-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Features in Eiav Ncp11: A Lentivirus Nucleocapsid Protein with a Short Linker
Biochemistry, 45, 2006
|
|
2BL7
 
 | | 1.6 Angstrom crystal structure of EntA-im: a bacterial immunity protein conferring immunity to the antimicrobial activity of the pediocin-like bacteriocin, enterocin A | | Descriptor: | ENTEROCINE A IMMUNITY PROTEIN | | Authors: | Johnsen, L, Dalhus, B, Leiros, I, Nissen-Meyer, J. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 1.6-A Crystal Structure of Enta-Im: A Bacterial Immunity Protein Conferring Immunity to the Antimicrobial Activity of the Pediocin-Like Bacteriocin Enterocin A
J.Biol.Chem., 280, 2005
|
|
2BL8
 
 | | 1.6 Angstrom crystal structure of EntA-im: a bacterial immunity protein conferring immunity to the antimicrobial activity of the pediocin-like bacteriocin, enterocin A | | Descriptor: | CITRATE ANION, ENTEROCINE A IMMUNITY PROTEIN | | Authors: | Johnsen, L, Dalhus, B, Leiros, I, Nissen-Meyer, J. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6-A Crystal Structure of Enta-Im: A Bacterial Immunity Protein Conferring Immunity to the Antimicrobial Activity of the Pediocin-Like Bacteriocin Enterocin A
J.Biol.Chem., 280, 2005
|
|
2BL9
 
 | | X-ray crystal structure of Plasmodium vivax dihydrofolate reductase in complex with pyrimethamine and its derivative | | Descriptor: | 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kongsaeree, P, Khongsuk, P, Leartsakulpanich, U, Chitnumsub, P, Tarnchompoo, B, Walkinshaw, M.D, Yuthavong, Y. | | Deposit date: | 2005-03-02 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Plasmodium Vivax: Pyrimethamine Displacement Linked with Mutation-Induced Resistance.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BLA
 
 | | SP21 double mutant P. vivax Dihydrofolate reductase in complex with pyrimethamine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE, ... | | Authors: | Kongsaeree, P, Khongsuk, P, Leartsakulpanich, U, Chitnumsub, P, Tarnchompoo, B, Walkinshaw, M.D, Yuthavong, Y. | | Deposit date: | 2005-03-02 | | Release date: | 2005-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Plasmodium Vivax: Pyrimethamine Displacement Linked with Mutation-Induced Resistance.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BLB
 
 | | X-ray crystal structure of Plasmodium vivax dihydrofolate reductase in complex with pyrimethamine and its derivative | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-ETHYL-5-PHENYLPYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE, ... | | Authors: | Kongsaeree, P, Khongsuk, P, Leartsakulpanich, U, Chitnumsub, P, Tarnchompoo, B, Walkinshaw, M.D, Yuthavong, Y. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Plasmodium Vivax: Pyrimethamine Displacement Linked with Mutation-Induced Resistance.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BLC
 
 | | SP21 double mutant P. vivax Dihydrofolate reductase in complex with des-chloropyrimethamine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-ETHYL-5-PHENYLPYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE, ... | | Authors: | Kongsaeree, P, Khongsuk, P, Leartsakulpanich, U, Chitnumsub, P, Tarnchompoo, B, Walkinshaw, M.D, Yuthavong, Y. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Plasmodium Vivax: Pyrimethamine Displacement Linked with Mutation-Induced Resistance.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BLE
 
 | | Structure of human guanosine monophosphate reductase GMPR1 in complex with GMP | | Descriptor: | GMP REDUCTASE I, GUANOSINE-5'-MONOPHOSPHATE, SODIUM ION | | Authors: | Bunkoczi, G, Haroniti, A, Ng, S, von Delft, F, Oppermann, U, Arrowsmith, C, Sundstrom, M, Edwards, A, Gileadi, O. | | Deposit date: | 2005-03-03 | | Release date: | 2005-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Human Guanosine Monophosphate Reductase Gmpr1 in Complex with Gmp
To be Published
|
|
2BLF
 
 | | Sulfite dehydrogenase from Starkeya Novella | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, HEME C, SULFATE ION, ... | | Authors: | Bailey, S, Kappler, U. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Intramolecular Electron Transfer in Sulfite-Oxidizing Enzymes is Revealed by High Resolution Structure of a Heterodimeric Complex of the Catalytic Molybdopterin Subunit and a C-Type Cytochrome Subunit.
J.Biol.Chem., 280, 2005
|
|
2BLG
 
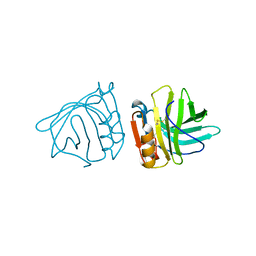 | | STRUCTURAL BASIS OF THE TANFORD TRANSITION OF BOVINE BETA-LACTOGLOBULIN FROM CRYSTAL STRUCTURES AT THREE PH VALUES; PH 8.2 | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Qin, B.Y, Bewley, M.C, Creamer, L.K, Baker, H.M, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1999-01-27 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis of the Tanford transition of bovine beta-lactoglobulin.
Biochemistry, 37, 1998
|
|
2BLH
 
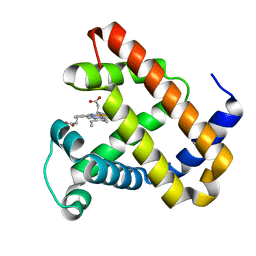 | | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
2BLI
 
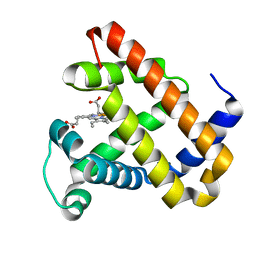 | | L29W Mb deoxy | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
2BLJ
 
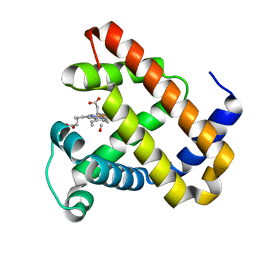 | | Structure of L29W MbCO | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
2BLL
 
 | | Apo-structure of the C-terminal decarboxylase domain of ArnA | | Descriptor: | PROTEIN YFBG | | Authors: | Williams, G.J, Breazeale, S.D, Raetz, C.R.H, Naismith, J.H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of Both Domains of Arna, a Dual Function Decarboxylase and a Formyltransferase, Involved in 4-Amino-4-Deoxy-L- Arabinose Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
2BLM
 
 | |
2BLN
 
 | | N-terminal formyltransferase domain of ArnA in complex with N-5- formyltetrahydrofolate and UMP | | Descriptor: | ACETATE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PROTEIN YFBG, ... | | Authors: | Williams, G.J, Breazeale, S.D, Raetz, C.R.H, Naismith, J.H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure and Function of Both Domains of Arna, a Dual Function Decarboxylase and a Formyltransferase, Involved in 4-Amino-4-Deoxy-L-Arabinose Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
2BLO
 
 | |
2BLP
 
 | | RNase before unattenuated X-RAY burn | | Descriptor: | CHLORIDE ION, RIBONUCLEASE PANCREATIC PRECURSOR | | Authors: | Nanao, M.H, Ravelli, R.B. | | Deposit date: | 2005-03-08 | | Release date: | 2005-09-07 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving radiation-damage substructures for RIP.
Acta Crystallogr. D Biol. Crystallogr., 61, 2005
|
|
2BLQ
 
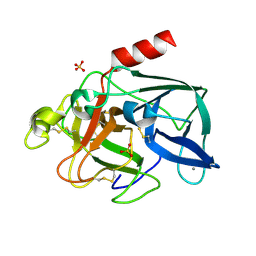 | | Elastase After A High Dose X-Ray "Burn" | | Descriptor: | CALCIUM ION, ELASTASE 1, SULFATE ION | | Authors: | Nanao, M.H, Ravelli, R.B. | | Deposit date: | 2005-03-08 | | Release date: | 2005-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Improving Radiation-Damage Substructures for Rip.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BLR
 
 | |
