2NWD
 
 | |
5EXN
 
 | | FACTOR XIA (C500S [C122S]) IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Authors: | Sheriff, S. | | Deposit date: | 2015-11-23 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Orally bioavailable pyridine and pyrimidine-based Factor XIa inhibitors: Discovery of the methyl N-phenyl carbamate P2 prime group
Bioorg.Med.Chem., 24, 2016
|
|
8PCH
 
 | | CRYSTAL STRUCTURE OF PORCINE CATHEPSIN H DETERMINED AT 2.1 ANGSTROM RESOLUTION: LOCATION OF THE MINI-CHAIN C-TERMINAL CARBOXYL GROUP DEFINES CATHEPSIN H AMINOPEPTIDASE FUNCTION | | Descriptor: | CATHEPSIN H, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guncar, G, Podobnik, M, Pungercar, J, Strukelj, B, Turk, V, Turk, D. | | Deposit date: | 1997-11-07 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of porcine cathepsin H determined at 2.1 A resolution: location of the mini-chain C-terminal carboxyl group defines cathepsin H aminopeptidase function.
Structure, 6, 1998
|
|
7VKV
 
 | |
3ADE
 
 | | Crystal Structure of Keap1 in Complex with Sequestosome-1/p62 | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION, Sequestosome-1 | | Authors: | Kurokawa, H, Yamamoto, M. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1
Nat.Cell Biol., 12, 2010
|
|
5I9U
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
7W11
 
 | | glycosyltransferase | | Descriptor: | DIGITOXIGENIN, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W0Z
 
 | | Glycosyltranferase UGT74AN2 | | Descriptor: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W09
 
 | | UGT74AN2, Plant Steroid Glycosyltransferase | | Descriptor: | Glycosyltransferase | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W1H
 
 | | glycosyltransferase | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W10
 
 | | UGT74AN2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, H, Long, F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W0K
 
 | | plant glycosyltransferase | | Descriptor: | GLYCEROL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W1B
 
 | | Glycosyltransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIGITOXIGENIN, Glycosyltransferase, ... | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
1ARZ
 
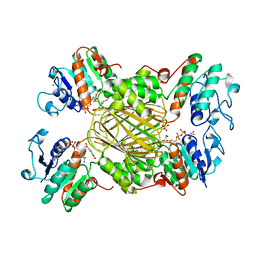 | | ESCHERICHIA COLI DIHYDRODIPICOLINATE REDUCTASE IN COMPLEX WITH NADH AND 2,6 PYRIDINE DICARBOXYLATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIHYDRODIPICOLINATE REDUCTASE, PHOSPHATE ION, ... | | Authors: | Scapin, G, Reddy, S.G, Zheng, R, Blanchard, J.S. | | Deposit date: | 1997-08-08 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of Escherichia coli dihydrodipicolinate reductase in complex with NADH and the inhibitor 2,6-pyridinedicarboxylate.
Biochemistry, 36, 1997
|
|
5IK1
 
 | |
6R6N
 
 | |
3X1H
 
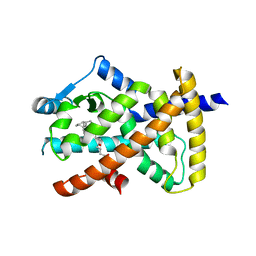 | | hPPARgamma Ligand binding domain in complex with 5-oxo-tricosahexaenoic acid | | Descriptor: | (7E,11Z,14Z,17Z,20Z)-5-oxotricosa-7,11,14,17,20-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-11-18 | | Release date: | 2015-04-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of covalent bond formation between PPAR gamma and oxo-fatty acids.
Bioconjug.Chem., 26, 2015
|
|
7WSB
 
 | | The ternary complex structure of FtmOx1 with a-ketoglutarate and 13-oxo-fumitremorgin B | | Descriptor: | 13-Oxofumitremorgin B, 2-OXOGLUTARIC ACID, COBALT (II) ION, ... | | Authors: | Wang, J, Wang, X.Y, Wang, Y.Y, Yan, W.P. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Dissecting the Mechanism of the Nonheme Iron Endoperoxidase FtmOx1 Using Substrate Analogues.
Jacs Au, 2, 2022
|
|
3X1I
 
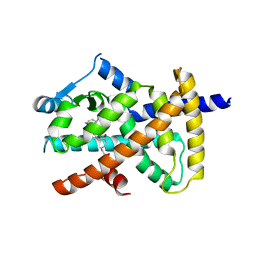 | | hPPARgamma Ligand binding domain in complex with 6-oxo-tetracosahexaenoic acid | | Descriptor: | (8E,12Z,15Z,18Z,21Z)-6-oxotetracosa-8,12,15,18,21-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-11-18 | | Release date: | 2015-04-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of covalent bond formation between PPAR gamma and oxo-fatty acids.
Bioconjug.Chem., 26, 2015
|
|
7QF6
 
 | |
5GIS
 
 | | Crystal structure of a Fab fragment with its ligand peptide | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, SULFATE ION, ... | | Authors: | Kitago, Y, Kaneko, K.K, Ogasawara, S, Kato, Y, Takagi, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for multi-specific peptide recognition by the anti-IDH1/2 monoclonal antibody, MsMab-1.
Biochem. Biophys. Res. Commun., 478, 2016
|
|
6ELB
 
 | | Tryptophan Repressor TrpR from E.coli variant M42F T44L T81M N87G S88Y with Indole-3-acetic acid as ligand | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Trp operon repressor | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-09-28 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.437 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
1GXM
 
 | | Family 10 polysaccharide lyase from Cellvibrio cellulosa | | Descriptor: | GLYCEROL, PECTATE LYASE | | Authors: | Charnock, S.J, Brown, I.E, Turkenburg, J.P, Black, G.W, Davies, G.J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Convergent Evolution Sheds Light on the Anti-Beta-Elimination Mechanism Common to Family 1 and 10 Polysaccharide Lyases
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1DBH
 
 | | DBL AND PLECKSTRIN HOMOLOGY DOMAINS FROM HSOS1 | | Descriptor: | PROTEIN (HUMAN SOS 1) | | Authors: | Soisson, S.M, Kuriyan, J. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Dbl and pleckstrin homology domains from the human Son of sevenless protein.
Cell(Cambridge,Mass.), 95, 1998
|
|
6RT2
 
 | | Crystal structure of Trypanosoma Brucei PEX14 N-terminal domain in complex with small molecules designed to investigate the water envelope | | Descriptor: | (3~{S})-3-[[1-(2-hydroxyethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-3-yl]carbonylamino]-3-phenyl-propanoic acid, BETA-MERCAPTOETHANOL, Peroxin 14, ... | | Authors: | Napolitano, V, Ratkova, E.L, Dawidowski, M, Dubin, G, Fino, R, Popowicz, G, Sattler, M, Tetko, I.V. | | Deposit date: | 2019-05-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Water envelope has a critical impact on the design of protein-protein interaction inhibitors.
Chem.Commun.(Camb.), 56, 2020
|
|
