1BCO
 
 | | BACTERIOPHAGE MU TRANSPOSASE CORE DOMAIN | | Descriptor: | BACTERIOPHAGE MU TRANSPOSASE | | Authors: | Rice, P.A, Mizuuchi, K. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration.
Cell(Cambridge,Mass.), 82, 1995
|
|
1AQM
 
 | | ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS COMPLEXED WITH TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 1997-07-31 | | Release date: | 1999-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor.
Protein Sci., 7, 1998
|
|
1AOA
 
 | | N-TERMINAL ACTIN-CROSSLINKING DOMAIN FROM HUMAN FIMBRIN | | Descriptor: | T-FIMBRIN | | Authors: | Goldsmith, S.C, Pokala, N, Shen, W, Fedorov, A.A, Matsudaira, P, Almo, S.C. | | Deposit date: | 1997-06-30 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of an actin-crosslinking domain from human fimbrin.
Nat.Struct.Biol., 4, 1997
|
|
1AQX
 
 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH MEISENHEIMER COMPLEX | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE | | Authors: | Prade, L, Huber, R, Manoharan, T.H, Fahl, W.E, Reuter, W. | | Deposit date: | 1997-08-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor.
Structure, 5, 1997
|
|
6ZT9
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
5EEW
 
 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 6.45 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1APA
 
 | | X-RAY STRUCTURE OF A POKEWEED ANTIVIRAL PROTEIN, CODED BY A NEW GENOMIC CLONE, AT 0.23 NM RESOLUTION. A MODEL STRUCTURE PROVIDES A SUITABLE ELECTROSTATIC FIELD FOR SUBSTRATE BINDING. | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Ago, H, Kataoka, J, Tsuge, H, Habuka, N, Inagaki, E, Noma, M, Miyano, M. | | Deposit date: | 1993-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of a pokeweed antiviral protein, coded by a new genomic clone, at 0.23 nm resolution. A model structure provides a suitable electrostatic field for substrate binding.
Eur.J.Biochem., 225, 1994
|
|
1APL
 
 | | CRYSTAL STRUCTURE OF A MAT-ALPHA2 HOMEODOMAIN-OPERATOR COMPLEX SUGGESTS A GENERAL MODEL FOR HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*C P*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT-ALPHA2 HOMEODOMAIN) | | Authors: | Wolberger, C, Vershon, A.K, Liu, B, Johnson, A.D, Pabo, C.O. | | Deposit date: | 1993-10-04 | | Release date: | 1993-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 67, 1991
|
|
5I0L
 
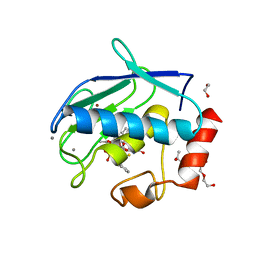 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated arylsulfonamide carboxylate water-soluble inhibitor (DC27). | | Descriptor: | (2R)-2-[{(E)-2-[({(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-bis(acetyloxy)-6-[(acetyloxy)methyl]tetrahydro-2H-pyran-2-yl}carbamothioyl)amino]ethenyl}(biphenyl-4-ylsulfonyl)amino]-3-methylbutanoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
6ZT7
 
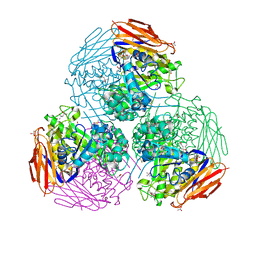 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
1AT6
 
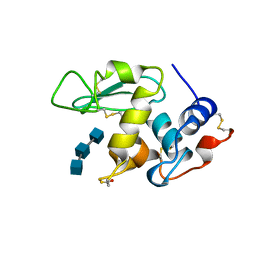 | | HEN EGG WHITE LYSOZYME WITH A ISOASPARTATE RESIDUE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Noguchi, S, Miyawaki, K, Satow, Y. | | Deposit date: | 1997-08-19 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Succinimide and isoaspartate residues in the crystal structures of hen egg-white lysozyme complexed with tri-N-acetylchitotriose.
J.Mol.Biol., 278, 1998
|
|
7ABI
 
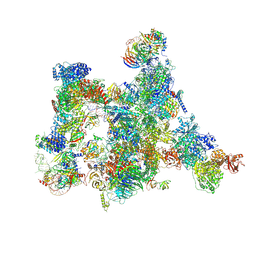 | | Human pre-Bact-2 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Beta-catenin-like protein 1, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
1ATL
 
 | | Structural interaction of natural and synthetic inhibitors with the VENOM METALLOPROTEINASE, ATROLYSIN C (FORM-D) | | Descriptor: | CALCIUM ION, O-methyl-N-[(2S)-4-methyl-2-(sulfanylmethyl)pentanoyl]-L-tyrosine, Snake venom metalloproteinase atrolysin-D, ... | | Authors: | Zhang, D, Botos, I, Gomis-Rueth, F.-X, Doll, R, Blood, C, Njoroge, F.G, Fox, J.W, Bode, W, Meyer, E.F. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d).
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
5EHD
 
 | | Crystal structure of human nucleophosmin-core in complex with cytochrome c | | Descriptor: | CHLORIDE ION, Nucleophosmin | | Authors: | Bernardo-Garcia, N, Hermoso, J.A, Gonzalez-Arzola, K, Diaz-Moreno, I, De la Rosa, M.A. | | Deposit date: | 2015-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of human nucleophosmin-core in complex with cytochrome c
To Be Published
|
|
1ASQ
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, AZIDE ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
5WOT
 
 | |
1AU7
 
 | | PIT-1 MUTANT/DNA COMPLEX | | Descriptor: | CONSENSUS DNA 25-MER, DNA (5'-D(*CP*TP*TP*CP*CP*TP*CP*AP*TP*GP*TP*AP*TP*AP*TP*AP*C P*AP*TP*GP*AP*GP* GP*A)-3'), PROTEIN PIT-1 | | Authors: | Jacobson, E.M, Li, P, Leon-Del-Rio, A, Rosenfeld, M.G, Aggarwal, A.K. | | Deposit date: | 1997-09-12 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility.
Genes Dev., 11, 1997
|
|
5X0S
 
 | | Solution NMR structure of peptide toxin SsTx from Scolopendra subspinipes mutilans | | Descriptor: | SsTx | | Authors: | Wu, F, Luo, L, Qu, D, Zhang, L, Tian, C, Lai, R. | | Deposit date: | 2017-01-23 | | Release date: | 2018-01-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Centipedes subdue giant prey by blocking KCNQ channels
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1AUR
 
 | |
5I2N
 
 | | Structure of the human GluN1/GluN2A LBD in complex with N-ethyl-7-{[2-fluoro-3-(trifluoromethyl)phenyl]methyl}-2-methyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 29) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
1AV4
 
 | |
1AVA
 
 | | AMY2/BASI PROTEIN-PROTEIN COMPLEX FROM BARLEY SEED | | Descriptor: | BARLEY ALPHA-AMYLASE 2(CV MENUET), BARLEY ALPHA-AMYLASE/SUBTILISIN INHIBITOR, CALCIUM ION | | Authors: | Vallee, F, Kadziola, A, Bourne, Y, Juy, M, Svensson, B, Haser, R. | | Deposit date: | 1997-09-15 | | Release date: | 1999-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Barley alpha-amylase bound to its endogenous protein inhibitor BASI: crystal structure of the complex at 1.9 A resolution.
Structure, 6, 1998
|
|
1AW1
 
 | |
1AVM
 
 | |
1AX0
 
 | |
