3JVB
 
 | |
3JVR
 
 | | Characterization of the Chk1 allosteric inhibitor binding site | | Descriptor: | (1S)-1-(1H-benzimidazol-2-yl)ethyl (3,4-dichlorophenyl)carbamate, Serine/threonine-protein kinase Chk1 | | Authors: | Chen, P. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
3VE2
 
 | |
3VES
 
 | | Crystal structure of the O-carbamoyltransferase TobZ in complex with AMPCPP and carbamoyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FE (II) ION, ... | | Authors: | Parthier, C, Stubbs, M.T, Goerlich, S, Jaenecke, F. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3J92
 
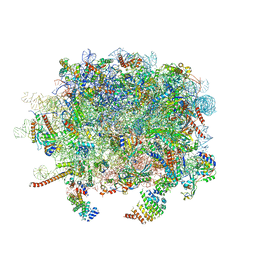 | | Structure and assembly pathway of the ribosome quality control complex | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Shao, S, Brown, A, Santhanam, B, Hegde, R.S. | | Deposit date: | 2014-12-02 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and Assembly Pathway of the Ribosome Quality Control Complex.
Mol.Cell, 57, 2015
|
|
3VL4
 
 | |
3JWT
 
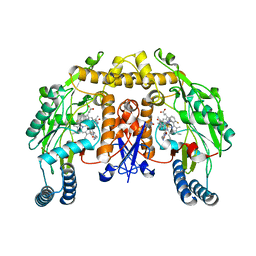 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with N1-{(3'R,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3VM0
 
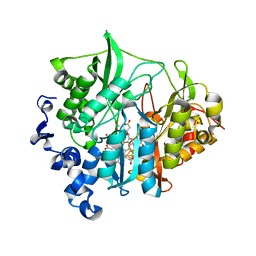 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - NO2 complex from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
3JX1
 
 | | Structure of rat neuronal nitric oxide synthase D597N mutant heme domain in complex with N1-{(3'R,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3VFI
 
 | | Crystal Structure of a Metagenomic Thioredoxin | | Descriptor: | thioredoxin | | Authors: | Craig, T.K, Gardberg, A, Lorimer, D.D, Burgin Jr, A.B, Segall, A, Rohwer, F. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a Metagenomic Thioredoxin
To be Published
|
|
3VFS
 
 | | crystal structure of HLA B*3508LPEP-P5Ala , peptide mutant P5-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P5A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3JYI
 
 | | Structural and biochemical evidence that a TEM-1 {beta}-lactamase Asn170Gly active site mutant acts via substrate-assisted catalysis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Brown, N.G, Palzkill, T.G, Prasad, B.V.V, Shanker, S. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis
J.Biol.Chem., 284, 2009
|
|
3VNT
 
 | | Crystal Structure of the Kinase domain of Human VEGFR2 with a [1,3]thiazolo[5,4-b]pyridine derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-3-(1-cyanocyclopropyl)-N-[5-({2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}oxy)-2-fluorophenyl]benzamide, Vascular endothelial growth factor receptor 2 | | Authors: | Oki, H. | | Deposit date: | 2012-01-17 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design and synthesis of novel DFG-out RAF/vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors. 1. Exploration of [5,6]-fused bicyclic scaffolds
J.Med.Chem., 55, 2012
|
|
3VG2
 
 | | Iodide derivative of human LFABP | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3K1G
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Ser-L-Tyr | | Descriptor: | Dipeptide Epimerase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VO3
 
 | | Crystal Structure of the Kinase domain of Human VEGFR2 with imidazo[1,2-b]pyridazine derivative | | Descriptor: | 1,2-ETHANEDIOL, N-[3-({2-[(cyclopropylcarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}oxy)phenyl]-1,3-dimethyl-1H-pyrazole-5-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Oki, H, Okada, K. | | Deposit date: | 2012-01-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of N-[5-({2-[(cyclopropylcarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}oxy)-2-methylphenyl]-1,3-dimethyl-1H-pyrazole-5-carboxamide (TAK-593), a highly potent VEGFR2 kinase inhibitor
Bioorg.Med.Chem., 21, 2013
|
|
3VOK
 
 | | X-ray Crystal Structure of Wild Type HrtR in the Apo Form with the Target DNA. | | Descriptor: | 5'-D(*AP*TP*GP*AP*CP*AP*CP*TP*GP*TP*GP*TP*CP*AP*T)-3', Transcriptional regulator | | Authors: | Sawai, H, Sugimoto, H, Shiro, Y, Aono, S. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Transcriptional Regulation of Heme Homeostasis in Lactococcus lactis.
J.Biol.Chem., 287, 2012
|
|
3K2T
 
 | | Crystal structure of Lmo2511 protein from Listeria monocytogenes, northeast structural genomics consortium target LkR84A | | Descriptor: | Lmo2511 protein | | Authors: | Seetharaman, J, Su, M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Lmo2511 protein from Listeria monocytogenes, northeast structural genomics consortium target LkR84A
To be Published
|
|
3VOX
 
 | |
3K2W
 
 | | CRYSTAL STRUCTURE OF betaine-aldehyde dehydrogenase FROM Pseudoalteromonas atlantica T6c | | Descriptor: | ACETATE ION, Betaine-aldehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF betaine-aldehyde dehydrogenase FROM Pseudoalteromonas atlantica
To be Published
|
|
3K44
 
 | | Crystal Structure of Drosophila melanogaster Pur-alpha | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Purine-rich binding protein-alpha, ... | | Authors: | Graebsch, A, Roche, S, Niessing, D. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of Pur-alpha reveals a Whirly-like fold and an unusual nucleic-acid binding surface
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K5D
 
 | | Crystal Structure of BACE-1 in complex with AHM178 | | Descriptor: | Beta-secretase 1, N-acetyl-L-leucyl-N-[(4S,5S,7R)-8-(butylamino)-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]-L-methioninamide | | Authors: | Rondeau, J.-M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design and synthesis of novel P2/P3 modified, non-peptidic beta-secretase (BACE-1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3VPO
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
3J1V
 
 | |
1W46
 
 | | P4 protein from Bacteriophage PHI12 in complex with ADP and MG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
