6UPN
 
 | | Endophilin B1 helical scaffold | | Descriptor: | Endophilin-B1 | | Authors: | Bhatt, V.S, Sundborger-Lunna, A.C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Amphipathic Motifs Regulate N-BAR Protein Endophilin B1 Auto-inhibition and Drive Membrane Remodeling.
Structure, 29, 2021
|
|
1CVX
 
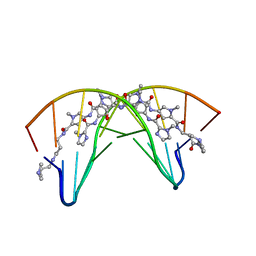 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYHPPYBETADP)2 BOUND TO B-DNA DECAMER CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', HYDROXYPYRROLE-IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1CW4
 
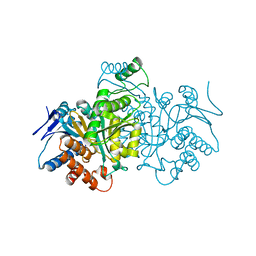 | |
1CWH
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH 3-D-SER CYCLOSPORIN | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-05-07 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | X-Ray Structures and Analysis of 11 Cyclosporin Derivatives Complexed with Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
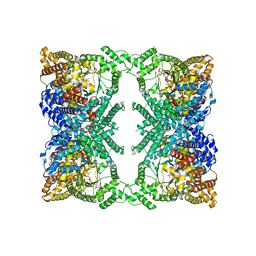
6V3O
 
 | |
1CZ3
 
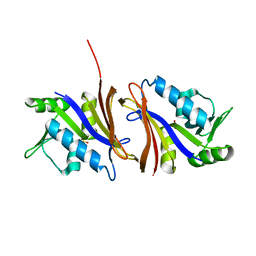 | | DIHYDROFOLATE REDUCTASE FROM THERMOTOGA MARITIMA | | Descriptor: | DIHYDROFOLATE REDUCTASE, SULFATE ION | | Authors: | Dams, T, Auerbach, G, Bader, G, Ploom, T, Huber, R, Jaenicke, R. | | Deposit date: | 1999-09-01 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of dihydrofolate reductase from Thermotoga maritima: molecular features of thermostability.
J.Mol.Biol., 297, 2000
|
|
8GN2
 
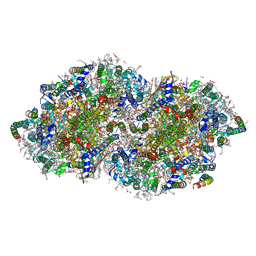 | | Crystal structure of PPBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
8GN0
 
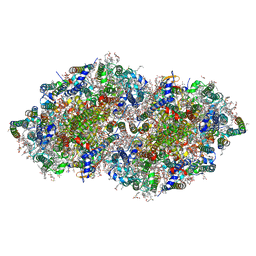 | | Crystal structure of DCBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
1D1G
 
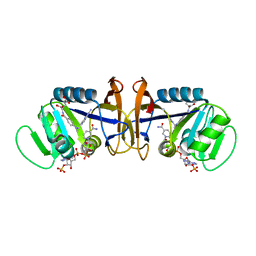 | | DIHYDROFOLATE REDUCTASE FROM THERMOTOGA MARITIMA | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dams, T, Auerbach, G, Bader, G, Ploom, T, Huber, R, Jaenicke, R. | | Deposit date: | 1999-09-16 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of dihydrofolate reductase from Thermotoga maritima: molecular features of thermostability.
J.Mol.Biol., 297, 2000
|
|
6V7B
 
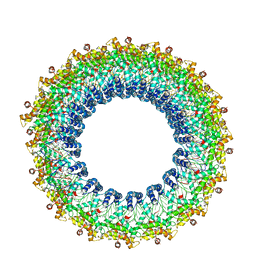 | | Cryo-EM reconstruction of Pyrobaculum filamentous virus 2 (PFV2) | | Descriptor: | A-DNA, Structural protein VP1, Structural protein VP2 | | Authors: | Wang, F, Baquero, D.P, Su, Z, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2019-12-08 | | Release date: | 2020-04-01 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a filamentous virus uncovers familial ties within the archaeal virosphere.
Virus Evol, 6, 2020
|
|
1D3V
 
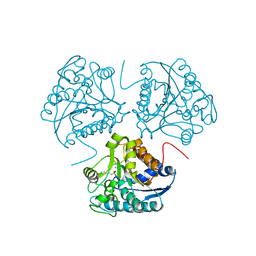 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH 2(S)-AMINO-6-BORONOHEXANOIC ACID, AN L-ARGININE ANALOG | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Cox, J.D, Kim, N.N, Traish, A.M, Christianson, D.W. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Arginase-boronic acid complex highlights a physiological role in erectile function.
Nat.Struct.Biol., 6, 1999
|
|
8JMN
 
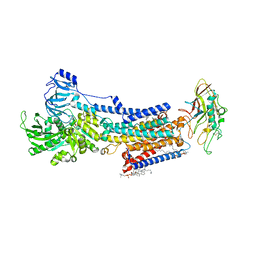 | | Cryo-EM structure of the gastric proton pump with bound DQ-21 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[[2-[(4-chlorophenyl)methoxy]phenyl]methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-06-05 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
1D4M
 
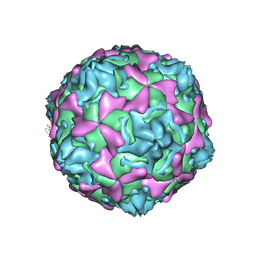 | | THE CRYSTAL STRUCTURE OF COXSACKIEVIRUS A9 TO 2.9 A RESOLUTION | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, PROTEIN (COXSACKIEVIRUS A9) | | Authors: | Hendry, E, Hatanaka, H, Fry, E, Smyth, M, Tate, J, Stanway, G, Santti, J, Maaronen, M, Hyypia, T, Stuart, D. | | Deposit date: | 1999-10-04 | | Release date: | 1999-12-23 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses.
Structure Fold.Des., 7, 1999
|
|
1D6O
 
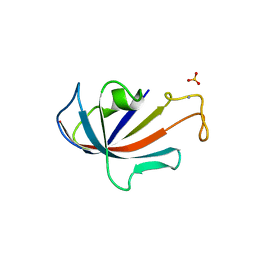 | | NATIVE FKBP | | Descriptor: | AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), SULFATE ION | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-15 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
8GJV
 
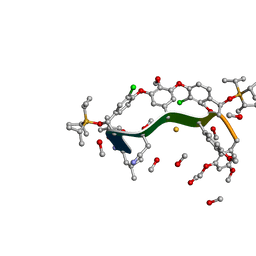 | |
1CP9
 
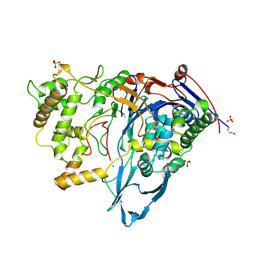 | |
1CQE
 
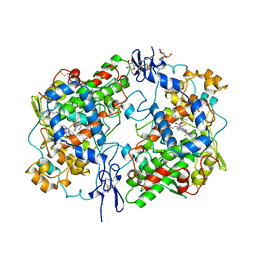 | | PROSTAGLANDIN H2 SYNTHASE-1 COMPLEX WITH FLURBIPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, PROTEIN (PROSTAGLANDIN H2 SYNTHASE-1), ... | | Authors: | Picot, D, Loll, P.J, Mulichak, A.M, Garavito, R.M. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1.
Nature, 367, 1994
|
|
1CR4
 
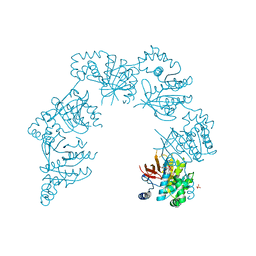 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTDP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CSQ
 
 | |
6VAT
 
 | |
8GTP
 
 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
1CTN
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL CHITINASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CHITINASE A | | Authors: | Perrakis, A, Tews, I, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bacterial chitinase at 2.3 A resolution.
Structure, 2, 1994
|
|
1CET
 
 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | Descriptor: | N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, PROTEIN (L-LACTATE DEHYDROGENASE) | | Authors: | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | Deposit date: | 1999-03-10 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
1CVN
 
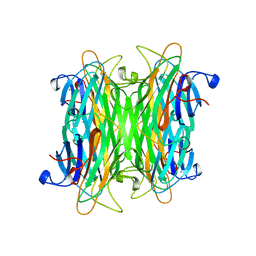 | | CONCANAVALIN A COMPLEXED TO TRIMANNOSIDE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Naismith, J.H. | | Deposit date: | 1995-08-09 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of trimannoside recognition by concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1CF5
 
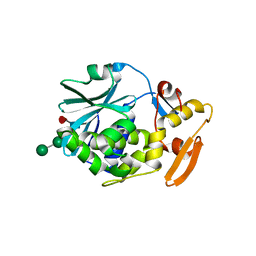 | | BETA-MOMORCHARIN STRUCTURE AT 2.55 A | | Descriptor: | PROTEIN (BETA-MOMORCHARIN), beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yuan, Y.-R, He, Y.-N, Xiong, J.-P, Xia, Z.-X. | | Deposit date: | 1999-03-24 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structure of beta-momorcharin at 2.55 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
