2R4I
 
 | |
4IR0
 
 | | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, FOSFOMYCIN, Metallothiol transferase FosB 2, ... | | Authors: | Maltseva, N, Kim, Y, Jedrzejczak, R, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames
To be Published
|
|
5D7W
 
 | | Crystal structure of serralysin | | Descriptor: | CALCIUM ION, GLYCEROL, Serralysin, ... | | Authors: | Wu, D, Ran, T, Xu, D.Q, Wang, W. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of a thermostable serralysin from Serratia sp. FS14 at 1.1 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6BDI
 
 | | Crystal structure of human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(1H-indol-3-yl)-3-{[(2R)-3-oxo-2-[(propan-2-yl)amino]-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-10-23 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Interaction of the rationally designed ritonavir-like inhibitors with human cytochrome P450 3A4: Impact of the side group interplay
Mol. Pharm., 2017
|
|
9BJS
 
 | | X-ray crystal structure of Y62W Thermothelomyces thermophilus polysaccharide monooxygenase 9E | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, COPPER (II) ION, ... | | Authors: | Thomas, W.C, Sayler, R.I, Marletta, M.A. | | Deposit date: | 2024-04-25 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Electron transfer in polysaccharide monooxygenase catalysis.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9BJR
 
 | | X-ray crystal structure of Y168F variant Thermothelomyces thermophilus polysaccharide monooxygenase 9E | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COPPER (II) ION, ... | | Authors: | Thomas, W.C, Sayler, R.I, Marletta, M.A. | | Deposit date: | 2024-04-25 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Electron transfer in polysaccharide monooxygenase catalysis.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6TWS
 
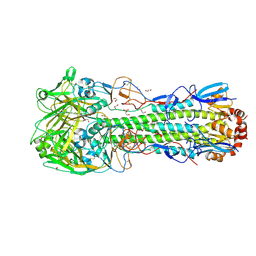 | | Crystal structure of the haemagglutinin mutant (Gln226Leu, Gly228Ser) from an H10N7 seal influenza virus isolated in Germany with 2mM EDTA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-13 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
9BJT
 
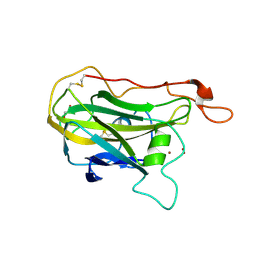 | | X-ray crystal structure of Y62F Thermothelomyces thermophilus polysaccharide monooxygenase 9E | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Glycoside hydrolase family 61 protein, ... | | Authors: | Thomas, W.C, Sayler, R.I, Marletta, M.A. | | Deposit date: | 2024-04-25 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Electron transfer in polysaccharide monooxygenase catalysis.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9BJQ
 
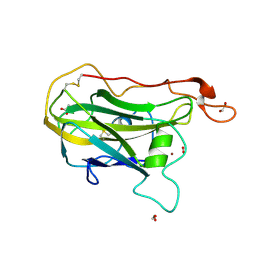 | | X-ray crystal structure of wild-type Thermothelomyces thermophilus polysaccharide monooxygenase 9E | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COPPER (II) ION, ... | | Authors: | Thomas, W.C, Sayler, R.I, Marletta, M.A. | | Deposit date: | 2024-04-25 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Electron transfer in polysaccharide monooxygenase catalysis.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6GJA
 
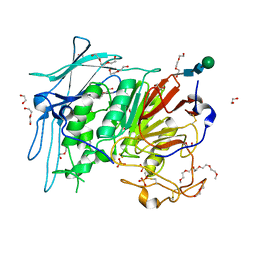 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - H229A MUTANT | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6ET0
 
 | | Crystal structure of PqsBC (C129A) mutant from Pseudomonas aeruginosa (crystal form 1) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Witzgall, F, Blankenfeldt, W. | | Deposit date: | 2017-10-25 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Alkylquinolone Repertoire of Pseudomonas aeruginosa is Linked to Structural Flexibility of the FabH-like 2-Heptyl-3-hydroxy-4(1H)-quinolone (PQS) Biosynthesis Enzyme PqsBC.
Chembiochem, 19, 2018
|
|
7C64
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in an open state (Form II) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CARBON DIOXIDE, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
8IAR
 
 | | Respiratory complex CIII2, focus-refined of type I, Wild type mouse under thermoneutral temperature | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
9C76
 
 | | LRRK2 Roc domain RP (Ras-pocket) complexed to Divarasib | | Descriptor: | 1-{(3S)-4-[(7M)-7-[6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl]-6-chloro-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}quinazolin-4-yl]-3-methylpiperazin-1-yl}propan-1-one, BROMIDE ION, FLUORIDE ION, ... | | Authors: | Zhu, L.Y, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2024-06-10 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | LRRK2 Roc domain RP (Ras-pocket) complexed to Divarasib
To Be Published
|
|
8IB7
 
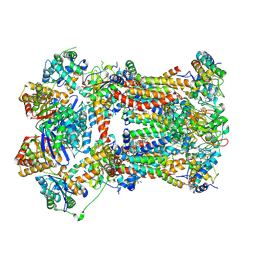 | | Respiratory complex CIII2, focus-refined of type IA, Wild type mouse under cold temperature | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
6BDM
 
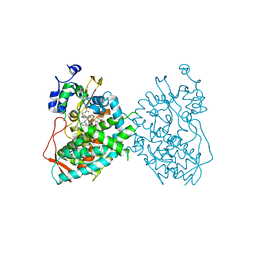 | | Crystal structure of human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(1H-indol-3-yl)-3-{[(2S)-3-oxo-2-(phenylamino)-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-10-23 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interaction of the rationally designed ritonavir-like inhibitors with human cytochrome P450 3A4: Impact of the side group interplay
Mol. Pharm., 2017
|
|
6QQY
 
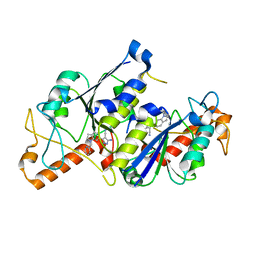 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-(pyridin-2-ylmethyl)indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6QR6
 
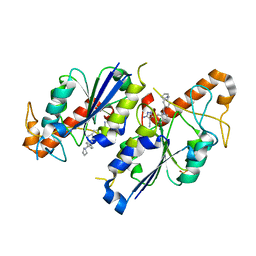 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
7FZD
 
 | | Crystal Structure of human FABP4 in complex with 3-(4-chlorophenyl)-4,5,6,6a-tetrahydro-3aH-cyclopenta[d][1,2]oxazole-5-carboxylic acid, i.e. SMILES [C@@H]12C(=NO[C@@H]1C[C@H](C2)C(=O)O)c1ccc(cc1)Cl with IC50=6.1 microM | | Descriptor: | (1R,3R,4S)-3-(4-chlorobenzoyl)-4-hydroxycyclopentane-1-carboxylic acid, (3aS,5R,6aS)-3-(4-chlorophenyl)-4,5,6,6a-tetrahydro-3aH-cyclopenta[d][1,2]oxazole-5-carboxylic acid, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Canesso, R, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
1XY6
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
7FWA
 
 | | Crystal Structure of human FABP4 in complex with 5-[(4-chloroanilino)methyl]-2-phenyl-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one, i.e. SMILES N12C(=NC(=N1)c1ccccc1)NC(=CC2=O)CNc1ccc(Cl)cc1 with IC50=0.683 microM | | Descriptor: | (8S)-5-[(4-chloroanilino)methyl]-2-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5YAL
 
 | | Ferulic acid esterase from Streptomyces cinnamoneus at 1.5 A resolution | | Descriptor: | ACETATE ION, Esterase, GLYCEROL, ... | | Authors: | Tamura, H, Uraji, M, Mizohata, E, Ogawa, K, Inoue, T, Hatanaka, T. | | Deposit date: | 2017-09-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Loop of Streptomyces Feruloyl Esterase Plays an Important Role in the Enzyme's Catalyzing the Release of Ferulic Acid from Biomass.
Appl. Environ. Microbiol., 84, 2018
|
|
4HGL
 
 | | Crystal structure of ck1g3 with compound 1 | | Descriptor: | 2-[5-methoxy-2-(quinolin-3-yl)pyrimidin-4-yl]-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Casein kinase I isoform gamma-3, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2012-10-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Potent and Selective CK1 gamma Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
6QQS
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 3-(1~{H}-indol-6-yl)-1~{H}-pyrazol-5-amine, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
9G9W
 
 | |
