7OZT
 
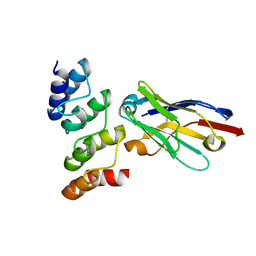 | | Nanobodies restore stability to cancer-associated mutants of tumor suppressor protein p16INK4a | | Descriptor: | Camelid nanobody NB09, Cyclin-dependent kinase inhibitor 2A | | Authors: | Pastok, M.W, Burbidge, O, Itzhaki, L, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Nanobodies restore stability to cancer-associated mutants of tumor suppressor protein p16INK4a
To Be Published
|
|
1HN3
 
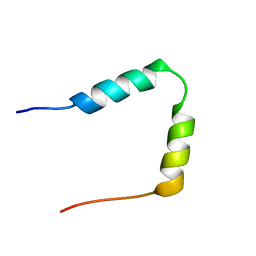 | | SOLUTION STRUCTURE OF THE N-TERMINAL 37 AMINO ACIDS OF THE MOUSE ARF TUMOR SUPPRESSOR PROTEIN | | Descriptor: | P19 ARF PROTEIN | | Authors: | DiGiammarino, E.L, Filippov, I, Weber, J.D, Bothner, B, Kriwacki, R.W. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the p53 regulatory domain of the p19Arf tumor suppressor protein.
Biochemistry, 40, 2001
|
|
2A5E
 
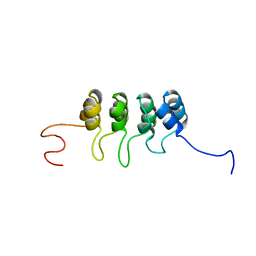 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, RESTRAINED MINIMIZED MEAN STRUCTURE | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
1A5E
 
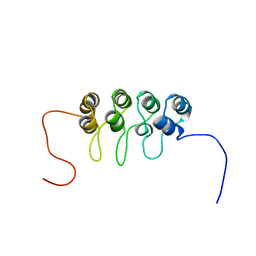 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 18 STRUCTURES | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
1BI7
 
 | | MECHANISM OF G1 CYCLIN DEPENDENT KINASE INHIBITION FROM THE STRUCTURE OF THE CDK6-P16INK4A TUMOR SUPPRESSOR COMPLEX | | Descriptor: | CYCLIN-DEPENDENT KINASE 6, MULTIPLE TUMOR SUPPRESSOR | | Authors: | Russo, A.A, Tong, L, Lee, J.O, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a.
Nature, 395, 1998
|
|
1DC2
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 20 STRUCTURES | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR A (P16INK4A) | | Authors: | Byeon, I.-J.L, Li, J, Yuan, C, Tsai, M.-D. | | Deposit date: | 1999-11-04 | | Release date: | 1999-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: refinement of p16INK4A structure and determination of p15INK4B structure by comparative modeling and NMR data.
Protein Sci., 9, 2000
|
|
