6W9T
 
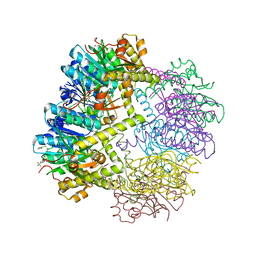 | |
6NAQ
 
 | | Crystal structure of Neisseria meningitidis ClpP protease in Apo form | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION | | Authors: | Houry, W.A, Mabanglo, M.F, Pai, E.F, Eger, B.T, Bryson, S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
6NAY
 
 | |
8SZN
 
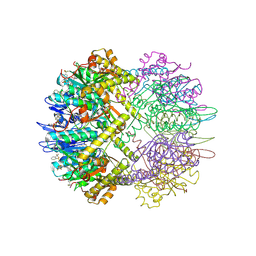 | |
5DKP
 
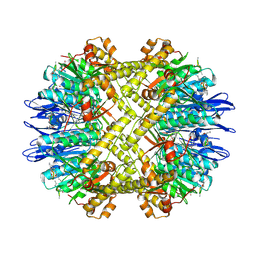 | | Crystal Structure of N. meningitidis ClpP in complex with agonist ADEP A54556. | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION, SODIUM ION, ... | | Authors: | Goodreid, J.D, Janetzko, J, Santa Maria Jr, J.P, Wong, K, Leung, E, Eger, B.T, Bryson, S, Pai, E.F, Gray-Owen, S.D, Walker, S, Houry, W.A, Batey, R.A. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Development and Characterization of Potent Cyclic Acyldepsipeptide Analogues with Increased Antimicrobial Activity.
J.Med.Chem., 59, 2016
|
|
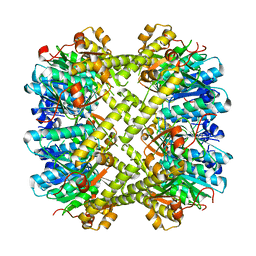
6NAW
 
 | |
6NAH
 
 | |
6VFX
 
 | | ClpXP from Neisseria meningitidis - Conformation B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
7KR2
 
 | |
6VFS
 
 | | ClpXP from Neisseria meningitidis - Conformation A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
