9D1Y
 
 | |
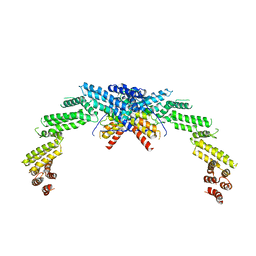
8W4J
 
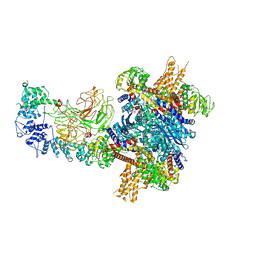 | |
8KHP
 
 | | CULLIN3-KLHL22-RBX1 E3 ligase | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch-like protein 22 | | Authors: | Su, M.-Y, Su, M.-Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-13 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
9D1Z
 
 | |
9D1I
 
 | |
5A10
 
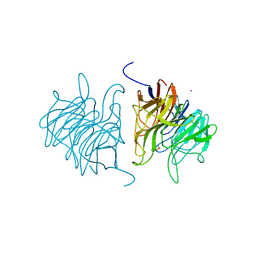 | | The crystal structure of Ta-TFP, a thiocyanate-forming protein involved in glucosinolate breakdown (space group C2) | | Descriptor: | ACETATE ION, SODIUM ION, THIOCYANATE FORMING PROTEIN | | Authors: | Krausze, J, Gumz, F, Wittstock, U. | | Deposit date: | 2015-04-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | The Crystal Structure of the Thiocyanate-Forming Protein from Thlaspi Arvense, a Kelch Protein Involved in Glucosinolate Breakdown.
Plant Mol.Biol., 89, 2015
|
|
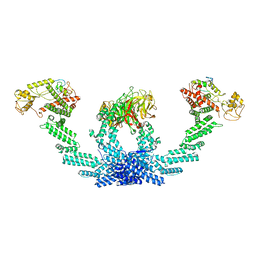
8K8T
 
 | | Structure of CUL3-RBX1-KLHL22 complex | | Descriptor: | Cullin-3, Kelch-like protein 22 | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
5A11
 
 | | The crystal structure of Ta-TFP, a thiocyanate-forming protein involved in glucosinolate breakdown (space group P21) | | Descriptor: | IODIDE ION, THIOCYANATE FORMING PROTEIN | | Authors: | Krausze, J, Gumz, F, Wittstock, U. | | Deposit date: | 2015-04-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The Crystal Structure of the Thiocyanate-Forming Protein from Thlaspi Arvense, a Kelch Protein Involved in Glucosinolate Breakdown.
Plant Mol.Biol., 89, 2015
|
|
