3ZHI
 
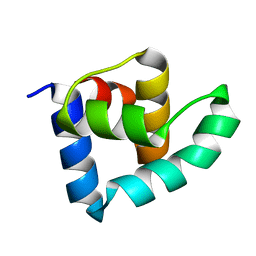 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 | | Descriptor: | CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
8QAO
 
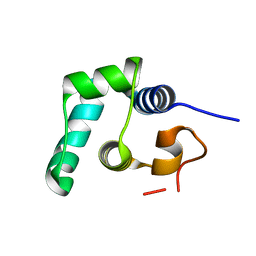 | |
3JXD
 
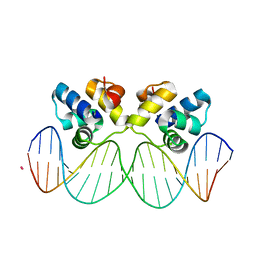 | |
1PRA
 
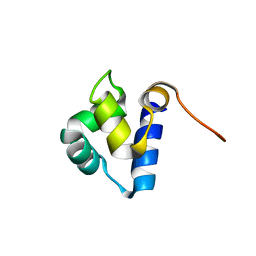 | |
2KPJ
 
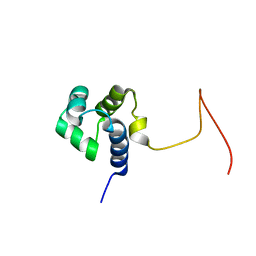 | | Solution Structure Of Protein SOS-response transcriptional repressor, LexA From Eubacterium rectale. Northeast Structural Genomics Consortium Target ErR9A | | Descriptor: | SOS-response transcriptional repressor, LexA | | Authors: | Wu, Y, Eletsky, A, Lee, D, Ghosh, A, Buchwald, W, Zhang, Q, Janjua, H, Garcia, E, Nair, R, Sukumaran, D, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-16 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of Protein SOS-response transcriptional repressor, LexA From Eubacterium rectale. Northeast Structural Genomics Consortium Target ErR9A
To be Published
|
|
7T8I
 
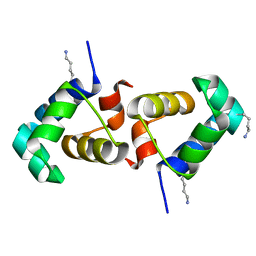 | |
5TN0
 
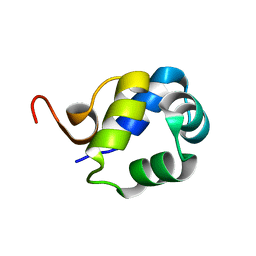 | | Solution Structure of the N-terminal DNA-binding domain of the master biofilm-regulator SinR from Bacillus subtilis | | Descriptor: | HTH-type transcriptional regulator SinR | | Authors: | Draughn, G.L, Bobay, B.G, Stowe, S.D, Thompson, R.J, Cavanagh, J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Solution Structures and Interaction of SinR and SinI: Elucidating the Mechanism of Action of the Master Regulator Switch for Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 2019
|
|
1LLI
 
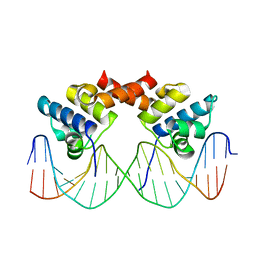 | | THE CRYSTAL STRUCTURE OF A MUTANT PROTEIN WITH ALTERED BUT IMPROVED HYDROPHOBIC CORE PACKING | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Lim, W.A, Hodel, A, Sauer, R.T, Richards, F.M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a mutant protein with altered but improved hydrophobic core packing.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
8BNY
 
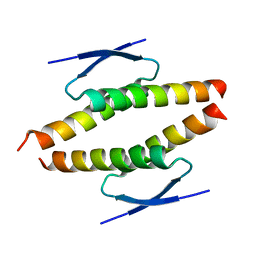 | | Structure of the tetramerization domain of pLS20 conjugation repressor Rco | | Descriptor: | CHLORIDE ION, Immunity repressor protein | | Authors: | Bernardo, N, Crespo, I, Meijer, W.J.J, Boer, D.R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | A tetramerization domain in prokaryotic and eukaryotic transcription regulators homologous to p53.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8DTQ
 
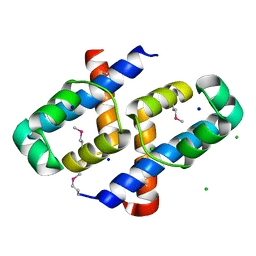 | | Crystal Structure of Staphylococcus aureus pSK41 Cop | | Descriptor: | CHLORIDE ION, Helix-turn-helix domain, SODIUM ION | | Authors: | Walton, W.G, Eakes, T.C, Redinbo, M.R, McLaughlin, K.J. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | pSK41/pGO1-family conjugative plasmids of Staphylococcus aureus encode a cryptic repressor of replication.
Plasmid, 128, 2023
|
|
1ADR
 
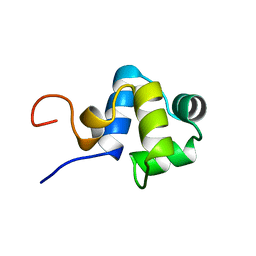 | |
8EZT
 
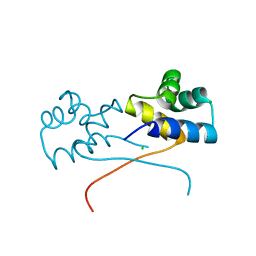 | | Crystal structure of HipB(Lp) from Legionella pneumophila | | Descriptor: | CHLORIDE ION, HipB(Lp) | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of HipB(Lp) from Legionella pneumophila
To Be Published
|
|
7ZVI
 
 | | Non-canonical Staphylococcus aureus pathogenicity island repression | | Descriptor: | Orf22, Sri | | Authors: | Miguel-Romero, L, Alqasmi, M, Bacarizo, J, Marina, A, Penades, J.R. | | Deposit date: | 2022-05-16 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.973 Å) | | Cite: | Non-canonical Staphylococcus aureus pathogenicity island repression.
Nucleic Acids Res., 50, 2022
|
|
1LMB
 
 | | REFINED 1.8 ANGSTROM CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Beamer, L.J, Pabo, C.O. | | Deposit date: | 1991-11-05 | | Release date: | 1991-11-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A crystal structure of the lambda repressor-operator complex.
J.Mol.Biol., 227, 1992
|
|
1LRP
 
 | |
6CHV
 
 | | Proteus vulgaris HigA antitoxin bound to DNA | | Descriptor: | Antitoxin HigA, MAGNESIUM ION, pHigCryst3, ... | | Authors: | Schureck, M.A, Hoffer, E.D, Onuoha, N, Dunham, C.M. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
7P4A
 
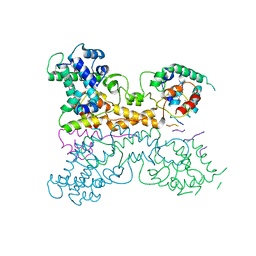 | | Non-canonical Staphylococcus aureus pathogenicity island repression. | | Descriptor: | Sri, Stl | | Authors: | Miguel-Romero, L, Alqasmi, M, Bacarizo, J, Tan, J.A, Cogdell, R.J, Chen, J, Byron, O, Christie, G.E, Marina, A, Penades, J.R. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Non-canonical Staphylococcus aureus pathogenicity island repression.
Nucleic Acids Res., 50, 2022
|
|
7N10
 
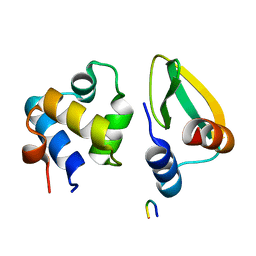 | |
1ZUG
 
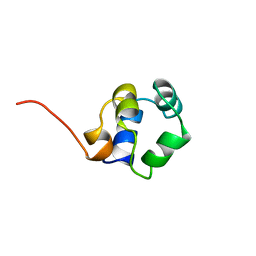 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | PHAGE 434 CRO PROTEIN | | Authors: | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | Deposit date: | 1997-03-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
7N1N
 
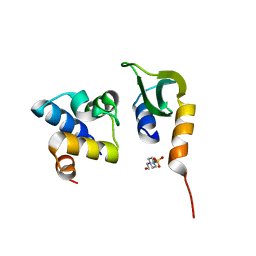 | | Prx in complex with ComR DNA-binding domain | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ComR, Prx | | Authors: | Rutbeek, N.R, Prehna, G. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of quorum sensing inhibition in Streptococcus by the phage protein paratox.
J.Biol.Chem., 297, 2021
|
|
5WOQ
 
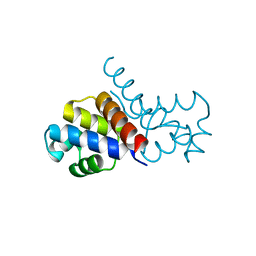 | |
6CF1
 
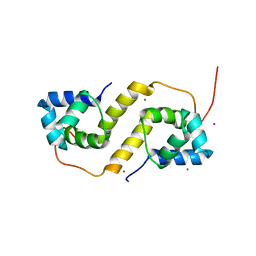 | | Proteus vulgaris HigA antitoxin structure | | Descriptor: | Antitoxin HigA, POTASSIUM ION | | Authors: | Schureck, M.A, Hoffer, E.D, Ei Cho, S, Dunham, C.M. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
1PER
 
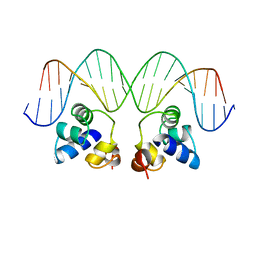 | |
4YG1
 
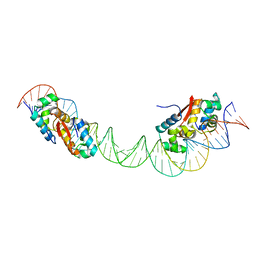 | | HipB-O1-O2 complex/P21212 crystal form | | Descriptor: | Antitoxin HipB, DNA (48-MER) | | Authors: | Schumacher, M.A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
1X57
 
 | | Solution structures of the HTH domain of human EDF-1 protein | | Descriptor: | Endothelial differentiation-related factor 1 | | Authors: | Nameki, N, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the HTH domain of human EDF-1 protein
To be Published
|
|
