3KHN
 
 | |
3IMP
 
 | |
5U1H
 
 | | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa | | Descriptor: | (2R,6S)-2-amino-6-(carboxyamino)-7-{[(1R)-1-carboxyethyl]amino}-7-oxoheptanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-28 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa
To be published
|
|
1R1M
 
 | |
5JIR
 
 | | Crystal structure of Treponema pallidum protein Tp0624 | | Descriptor: | CHLORIDE ION, GLYCEROL, OOP family OmpA-OmpF porin | | Authors: | Parker, M.L, Boulanger, M.J. | | Deposit date: | 2016-04-22 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Treponema pallidum Tp0624 Reveals a Modular Assembly of Divergently Functionalized and Previously Uncharacterized Domains.
Plos One, 11, 2016
|
|
7BBA
 
 | |
7RJJ
 
 | | Crystal Structure of the Peptidoglycan Binding Domain of the Outer Membrane Protein (OmpA) from Klebsiella pneumoniae with bound D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, OmpA family protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6IKJ
 
 | | Crystal structure of YfiB(F48S) | | Descriptor: | GLYCEROL, SULFATE ION, YfiB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis of activating mutants of YfiB from Pseudomonas aeruginosa PAO1.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6U83
 
 | | OmpA-like domain of FopA1 from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-ALANINE, Outer membrane associated protein, ... | | Authors: | Michalska, K, Skarina, T, Stogios, P.J, Di Leo, R, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-04 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3566 Å) | | Cite: | OmpA-like domain of FopA1 from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
1OAP
 
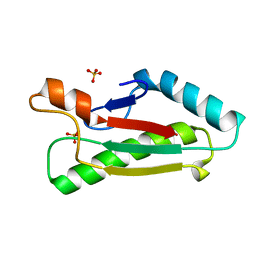 | | Mad structure of the periplasmique domain of the Escherichia coli PAL protein | | Descriptor: | PEPTIDOGLYCAN-ASSOCIATED LIPOPROTEIN, SULFATE ION | | Authors: | Abergel, C, Walburger, A, Bouveret, E, Claverie, J.M. | | Deposit date: | 2003-01-20 | | Release date: | 2004-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallization and preliminary crystallographic study of the peptidoglycan-associated lipoprotein from Escherichia coli.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2AIZ
 
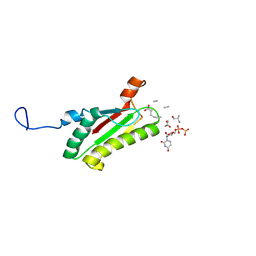 | | Solution structure of peptidoglycan associated lipoprotein from Haemophilus influenza bound to UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine | | Descriptor: | L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine, N-acetyl-beta-muramic acid, Outer membrane protein P6 (Fragment), ... | | Authors: | Parsons, L.M, Lin, F, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2005-08-01 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Peptidoglycan recognition by pal, an outer membrane lipoprotein.
Biochemistry, 45, 2006
|
|
9LJK
 
 | |
9LJL
 
 | |
9LJM
 
 | |
3CYQ
 
 | |
3LDT
 
 | |
4B62
 
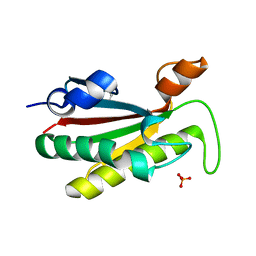 | | The structure of the cell wall anchor of the T6SS from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, TSSL1 | | Authors: | Robb, C.S, Carlson, M, Nano, F.E, Boraston, A.B. | | Deposit date: | 2012-08-08 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the Periplasmic Peptidoglycan Binding Anchor of a T6Ss from Pseudomonas Aeruginosa.
To be Published
|
|
4B5C
 
 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia pseudomallei | | Descriptor: | ACETATE ION, PUTATIVE OMPA FAMILY LIPOPROTEIN | | Authors: | Gourlay, L.J, Peri, C, Conchillo-Sole, O, Ferrer-Navarro, M, Gori, A, Longhi, R, Rinchai, D, Lertmemongkolchai, G, Lassaux, P, Daura, X, Colombo, G, Bolognesi, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Burkholderia Pseudomallei Acute Phase Antigen Bpsl2765 for Structure-Based Epitope Discovery/Design in Structural Vaccinology.
Chem.Biool., 20, 2013
|
|
3CYP
 
 | |
8VVN
 
 | |
4PWT
 
 | | Crystal structure of peptidoglycan-associated outer membrane lipoprotein from Yersinia pestis CO92 | | Descriptor: | FORMIC ACID, PYROPHOSPHATE 2-, Peptidoglycan-associated lipoprotein, ... | | Authors: | Maltseva, N, Kim, Y, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystal structure of peptidoglycan-associated outer membrane lipoprotein from
Yersinia pestis CO92
To be Published
|
|
6IKI
 
 | | Crystal structure of YfiB(W55L) | | Descriptor: | GLYCEROL, SULFATE ION, YfiB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural analysis of activating mutants of YfiB from Pseudomonas aeruginosa PAO1.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
4RHA
 
 | | Structure of the C-terminal domain of outer-membrane protein OmpA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Outer membrane protein A, ... | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-01 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
4ZHV
 
 | | Crystal structure of a bacterial signalling protein | | Descriptor: | SULFATE ION, YfiB | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
2K1S
 
 | | Solution NMR structure of the folded C-terminal fragment of YiaD from Escherichia coli. Northeast Structural Genomics Consortium target ER553. | | Descriptor: | Inner membrane lipoprotein yiaD | | Authors: | Ramelot, T.A, Zhao, L, Hamilton, K, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the folded C-terminal fragment of YiaD from Escherichia coli. Northeast Structural Genomics Consortium target ER553.
To be Published
|
|
