2LSO
 
 | | Solution NMR Structure of the Globular Domain of Human Histone H1x, Northeast Structural Genomics Consortium (NESG) Target HR7057A | | Descriptor: | Histone H1x | | Authors: | Eletsky, A, Lee, H, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-03 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Globular Domain of Human Histone H1x, Northeast Structural Genomics Consortium (NESG) Target HR7057A
To be Published
|
|
2RQP
 
 | |
1YQA
 
 | |
1GHC
 
 | | HOMO-AND HETERONUCLEAR TWO-DIMENSIONAL NMR STUDIES OF THE GLOBULAR DOMAIN OF HISTONE H1: FULL ASSIGNMENT, TERTIARY STRUCTURE, AND COMPARISON WITH THE GLOBULAR DOMAIN OF HISTONE H5 | | Descriptor: | GH1 | | Authors: | Cerf, C, Lippens, G, Ramakrishnan, V, Muyldermans, S, Segers, A, Wyns, L, Wodak, S.J, Hallenga, K. | | Deposit date: | 1994-05-16 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: full assignment, tertiary structure, and comparison with the globular domain of histone H5.
Biochemistry, 33, 1994
|
|
1HST
 
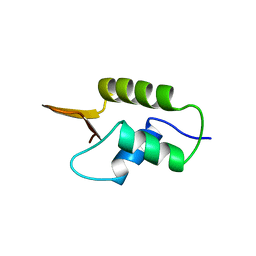 | | CRYSTAL STRUCTURE OF GLOBULAR DOMAIN OF HISTONE H5 AND ITS IMPLICATIONS FOR NUCLEOSOME BINDING | | Descriptor: | HISTONE H5 | | Authors: | Ramakrishnan, V, Finch, J.T, Graziano, V, Lee, P.L, Sweet, R.M. | | Deposit date: | 1993-03-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of globular domain of histone H5 and its implications for nucleosome binding.
Nature, 362, 1993
|
|
1UHM
 
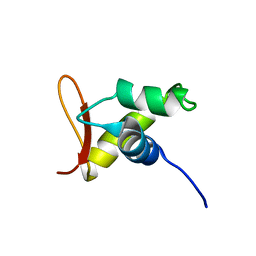 | | Solution structure of the globular domain of linker histone homolog Hho1p from S. cerevisiae | | Descriptor: | Histone H1 | | Authors: | Ono, K, Kusano, O, Shimotakahara, S, Shimizu, M, Yamazaki, T, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-05 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The linker histone homolog Hho1p from Saccharomyces cerevisiae represents a winged helix-turn-helix fold as determined by NMR spectroscopy.
Nucleic Acids Res., 31, 2003
|
|
1UST
 
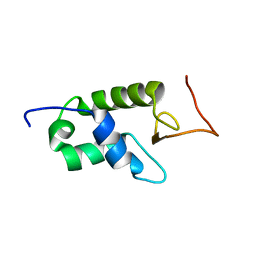 | | YEAST HISTONE H1 GLOBULAR DOMAIN I, HHO1P GI, SOLUTION NMR STRUCTURES | | Descriptor: | HISTONE H1 | | Authors: | Ali, T, Coles, P, Stevens, T.J, Stott, K, Thomas, J.O. | | Deposit date: | 2003-11-30 | | Release date: | 2004-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Homologous Domains of Similar Structure But Different Stability in the Yeast Linker Histone, Hho1P
J.Mol.Biol., 338, 2004
|
|
1USS
 
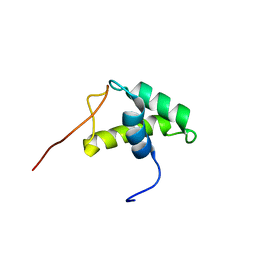 | | YEAST HISTONE H1 GLOBULAR DOMAIN II, HHO1P GII, SOLUTION NMR STRUCTURES | | Descriptor: | HISTONE H1 | | Authors: | Ali, T, Coles, P, Stevens, T.J, Stott, K, Thomas, J.O. | | Deposit date: | 2003-11-30 | | Release date: | 2004-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Homologous Domains of Similar Structure But Different Stability in the Yeast Linker Histone, Hho1P
J.Mol.Biol., 338, 2004
|
|
6HQ1
 
 | |
7C0J
 
 | | Crystal structure of chimeric mutant of GH5 in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Histone H5,Double-stranded RNA-specific adenosine deaminase | | Authors: | Choi, H.J, Park, C.H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
8VG2
 
 | |
5NL0
 
 | | Crystal structure of a 197-bp palindromic 601L nucleosome in complex with linker histone H1 | | Descriptor: | DNA (197-MER), Histone H1.0-B, Histone H2A type 1, ... | | Authors: | Garcia-Saez, I, Petosa, C, Dimitrov, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.4 Å) | | Cite: | Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.
Mol. Cell, 66, 2017
|
|
4QLC
 
 | | Crystal structure of chromatosome at 3.5 angstrom resolution | | Descriptor: | CITRIC ACID, DNA (167-mer), H5, ... | | Authors: | Jiang, J.S, Zhou, B.R, Xiao, T.S, Bai, Y.W. | | Deposit date: | 2014-06-11 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural Mechanisms of Nucleosome Recognition by Linker Histones.
Mol.Cell, 33 Suppl 1, 2015
|
|
8TB9
 
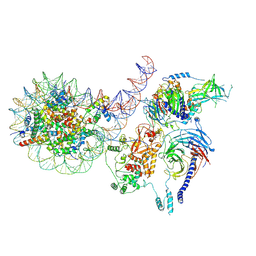 | | PRC2-J119-450 monomer bound to H1-nucleosome | | Descriptor: | DNA (226-MER), Histone H1.0, Histone H2A type 1, ... | | Authors: | Sauer, P.V, Cookis, T, Pavlenko, E, Nogales, E, Poepsel, S. | | Deposit date: | 2023-06-28 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of automethylated PRC2 by dimerization on chromatin.
Mol.Cell, 84, 2024
|
|
5WCU
 
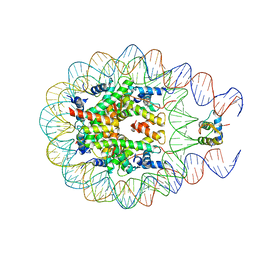 | |
9DDE
 
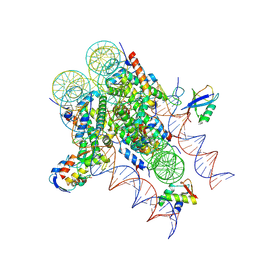 | | ncPRC1RYBP bound to H2AK119Ub/H1.4 chromatosome | | Descriptor: | DNA (187-MER), E3 ubiquitin-protein ligase RING2, Histone H1.4, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-08-28 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
8KE0
 
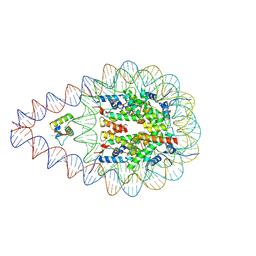 | | Structure of H1.2 bound to the nucleosome | | Descriptor: | DNA (193-MER), Histone H1.2, Histone H2A type 1-B/E, ... | | Authors: | Kujirai, T, Echigoya, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-08-11 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into how DEK nucleosome binding facilitates H3K27 trimethylation in chromatin.
Nat.Struct.Mol.Biol., 2025
|
|
7DBP
 
 | | Linker histone defines structure and self-association behaviour of the 177 bp human chromosome | | Descriptor: | DNA (175-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Wang, S, Vogirala, K.V, Soman, A, Liu, Z.B. | | Deposit date: | 2020-10-21 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Linker histone defines structure and self-association behaviour of the 177 bp human chromatosome.
Sci Rep, 11, 2021
|
|
6N88
 
 | |
9IPU
 
 | | cryo-EM structure of the RNF168(1-193)/UbcH5c-Ub ubiquitylation module bound to H1.0-K63-Ub3 modified chromatosome | | Descriptor: | DNA (171-MER), E3 ubiquitin-protein ligase RNF168, Histone H1.0, ... | | Authors: | Ai, H.S, Deng, Z.H, Liu, L. | | Deposit date: | 2024-07-11 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Promotion of RNF168-Mediated Nucleosomal H2A Ubiquitylation by Structurally Defined K63-Polyubiquitylated Linker Histone H1.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8XJV
 
 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
9J8O
 
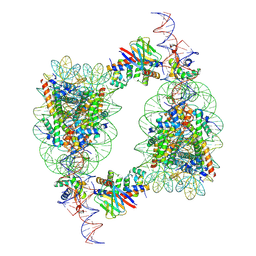 | | Cryo-EM structure of BAF-Lamin A/C IgF-H1-nucleosome complex | | Descriptor: | Barrier-to-autointegration factor, DNA (193-MER), Histone H1.1, ... | | Authors: | Horikoshi, N, Miyake, R, Sogawa-Fujiwara, C, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM structures of the BAF-Lamin A/C complex bound to nucleosomes.
Nat Commun, 16, 2025
|
|
6N89
 
 | |
8YTI
 
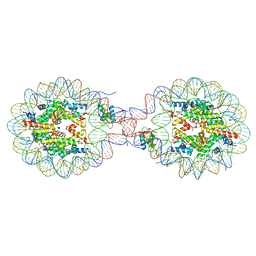 | | Crystal Structure of Nucleosome-H1x Linker Histone Assembly (sticky-169a DNA fragment) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Qiuye, B, Padavattan, S, Davey, C.A. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Linker Histones Associate Heterogeneously with Nucleosomes in the Condensed State
To Be Published
|
|
7COW
 
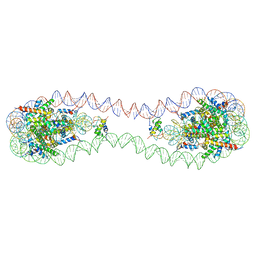 | | 353 bp di-nucleosome harboring cohesive DNA termini with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (353-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-08-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
