3P1T
 
 | |
3PA9
 
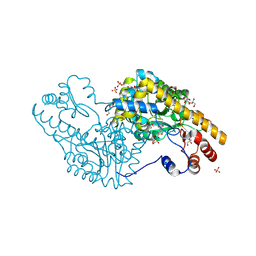 | | Mechanism of inactivation of E. coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) pH 7.5 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminofuran-2-carboxylic acid, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Fu, M, Silverman, R.B, Ringe, D. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
Biochemistry, 49, 2010
|
|
3QGU
 
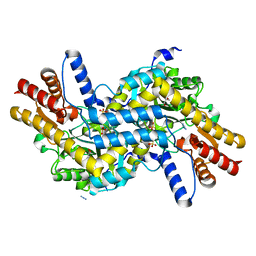 | | L,L-Diaminopimelate aminotransferase from Chlamydomonas reinhardtii | | Descriptor: | AZIDE ION, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Dobson, R.C.J, Giron, I, Hudson, A.O. | | Deposit date: | 2011-01-25 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | L,L-Diaminopimelate Aminotransferase from Chlamydomonas reinhardtii: A Target for Algaecide Development
Plos One, 6, 2011
|
|
3PIU
 
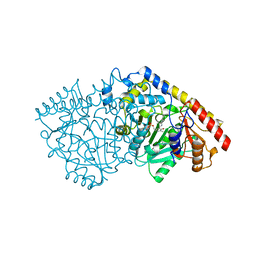 | |
3PD6
 
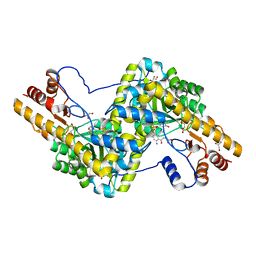 | | Crystal structure of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, ... | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV.
Biosci.Rep., 31, 2011
|
|
3PDB
 
 | | Crystal structure of mouse mitochondrial aspartate aminotransferase in complex with oxaloacetic acid | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, mitochondrial, ... | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV.
Biosci.Rep., 31, 2011
|
|
1IAX
 
 | | CRYSTAL STRUCTURE OF ACC SYNTHASE COMPLEXED WITH PLP | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Huai, Q, Xia, Y, Chen, Y, Callahan, B, Li, N, Ke, H. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5'-phosphate provide new insight into catalytic mechanisms
J.Biol.Chem., 276, 2001
|
|
3QPG
 
 | |
3QN6
 
 | |
1IJI
 
 | | Crystal Structure of L-Histidinol Phosphate Aminotransferase with PLP | | Descriptor: | Histidinol Phosphate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Li, Y, Larocque, R, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-29 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
3PDX
 
 | | Crystal structural of mouse tyrosine aminotransferase | | Descriptor: | Tyrosine aminotransferase | | Authors: | Mehere, P.V, Han, Q, Lemkul, J.A, Robinson, H, Bevan, D.R, Li, J. | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations.
Protein Cell, 1, 2010
|
|
1LKC
 
 | | Crystal Structure of L-Threonine-O-3-Phosphate Decarboxylase from Salmonella enterica | | Descriptor: | 1,2-ETHANEDIOL, L-threonine-O-3-phosphate decarboxylase, PHOSPHATE ION, ... | | Authors: | Cheong, C.G, Bauer, C.B, Brushaber, K.R, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2002-04-24 | | Release date: | 2002-05-01 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica.
Biochemistry, 41, 2002
|
|
1LC7
 
 | | Crystal Structure of L-Threonine-O-3-phosphate Decarboxylase from S. enterica complexed with a substrate | | Descriptor: | L-Threonine-O-3-Phosphate Decarboxylase, PHOSPHATE ION, PHOSPHOTHREONINE | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-04-05 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica: the apo, substrate, and product-aldimine complexes.
Biochemistry, 41, 2002
|
|
1IX8
 
 | | Aspartate Aminotransferase Active Site Mutant V39F/N194A | | Descriptor: | Aspartate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
1IX7
 
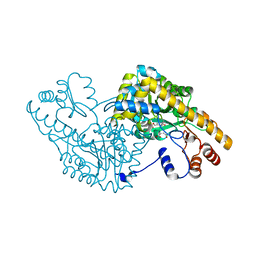 | | Aspartate Aminotransferase Active Site Mutant V39F maleate complex | | Descriptor: | Aspartate Aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
1LC5
 
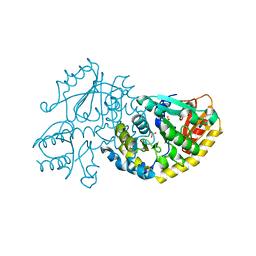 | | Crystal Structure of L-Threonine-O-3-phosphate Decarboxylase from S. enterica in its apo state | | Descriptor: | L-Threonine-O-3-Phosphate Decarboxylase, PHOSPHATE ION | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-04-05 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural studies of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica: the apo, substrate, and product-aldimine complexes.
Biochemistry, 41, 2002
|
|
6C3A
 
 | | O2-, PLP-dependent L-arginine hydroxylase RohP 4-hydroxy-2-ketoarginine complex | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
6C3C
 
 | | PLP-dependent L-arginine hydroxylase RohP quinonoid I complex | | Descriptor: | (2E)-5-carbamimidamido-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pentanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
6D0A
 
 | |
6DND
 
 | | Crystal structure of wild-type (WT) human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6DNB
 
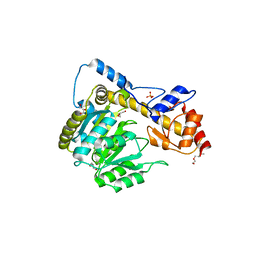 | | Crystal structure of T110A:S256A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, GLYCEROL, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6C9B
 
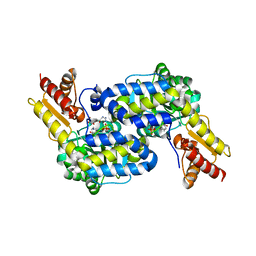 | | The structure of MppP soaked with the products 4HKA and 2KA | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, CHLORIDE ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-26 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
6C92
 
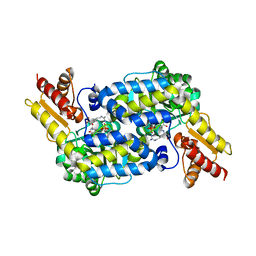 | | The structure of MppP soaked with the product 2-ketoarginine | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
6DNA
 
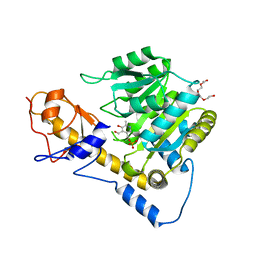 | | Crystal structure of T110A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6EZL
 
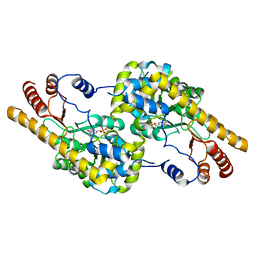 | |
