5FO5
 
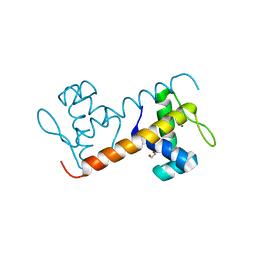 | | Structure of the DNA-binding domain of Escherichia coli methionine biosynthesis regulator MetR | | Descriptor: | 1,2-ETHANEDIOL, HTH-TYPE TRANSCRIPTIONAL REGULATOR METR, MAGNESIUM ION | | Authors: | Punekar, A.S, Porter, J, Urbanowski, M.L, Stauffer, G.V, Carr, S.B, Phillips, S.E. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for DNA Recognition by the Transcription Regulator Metr.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7TRV
 
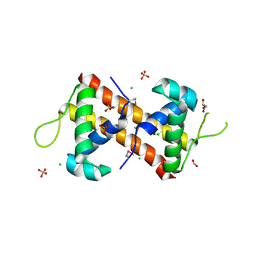 | | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Tesar, C, Crawford, M, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-31 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
4PZJ
 
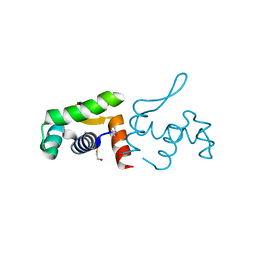 | | 1.60 Angstrom resolution crystal structure of a transcriptional regulator of the LysR family from Eggerthella lenta DSM 2243 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, LysR family | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Kiryukhina, O, Endres, M, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-31 | | Release date: | 2014-04-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of a transcriptional regulator of the LysR family from Eggerthella lenta DSM 2243
To be Published
|
|
5XXP
 
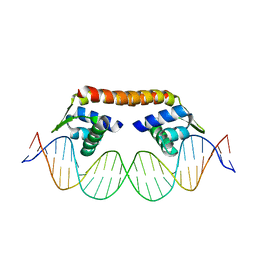 | | Crystal structure of CbnR_DBD-DNA complex | | Descriptor: | DNA (25-MER), LysR-type regulatory protein | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the DNA-binding domain of the LysR-type transcriptional regulator CbnR in complex with a DNA fragment of the recognition-binding site in the promoter region
FEBS J., 285, 2018
|
|
4IHS
 
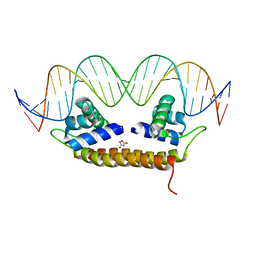 | | Crystal Structure of BenM_DBD/catB site 1 DNA Complex | | Descriptor: | HTH-type transcriptional regulator BenM, MALONATE ION, SODIUM ION, ... | | Authors: | Alanazi, A, Momany, C, Neidle, E.L. | | Deposit date: | 2012-12-19 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8WPA
 
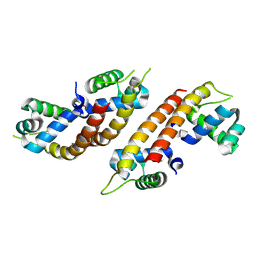 | |
4IHT
 
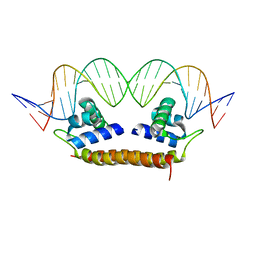 | | Crystal Structure of BenM_DBD/benA site 1 DNA Complex | | Descriptor: | HTH-type transcriptional regulator BenM, benA site 1 DNA, benA site 1 DNA - complement | | Authors: | Alanazi, A, Momany, C, Neidle, E.L. | | Deposit date: | 2012-12-19 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5Z4Y
 
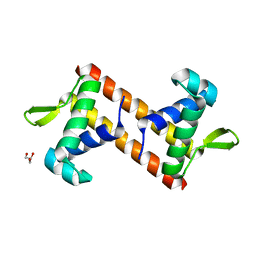 | |
3M1E
 
 | | Crystal Structure of BenM_DBD | | Descriptor: | HTH-type transcriptional regulator benM, SODIUM ION | | Authors: | Alanazi, A, Momany, C, Neidle, E.L. | | Deposit date: | 2010-03-04 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the unbound LysR-type transcriptional regulator BenM DNA binding domain
Acta Crystallogr.,Sect.D, 2013
|
|
9IRZ
 
 | | Crystal structure of YhaJ DNA-binding domain | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Probable HTH-type transcriptional regulator YhaJ | | Authors: | Kim, M, Kang, R, Ryu, S.E. | | Deposit date: | 2024-07-16 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | High-affinity promotor binding of YhaJ mediates a low signal leakage for effective DNT detection.
Front Microbiol, 15, 2024
|
|
6M5F
 
 | | Crystal structure of HinK, a LysR family transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator | | Authors: | Wang, Y, Lan, L, Cao, Q, Gan, J, Wang, F. | | Deposit date: | 2020-03-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of HinK, a LysR family transcriptional regulator from Pseudomonas aeruginosa
To Be Published
|
|
7L4S
 
 | |
7YHJ
 
 | |
4X6G
 
 | |
8SBF
 
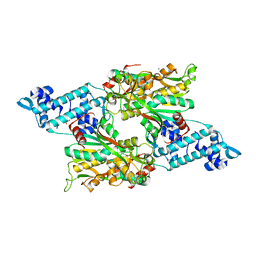 | | Full-length structure of the LysR-type transcriptional regulator, ACIAD0746, from Acinetobacter baylyi | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative transcriptional regulator (LysR family), ... | | Authors: | Momany, C, Nune, M, Brondani, J.C, Afful, D, Neidle, E, Galloway, N.R. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | FinR, a LysR-type transcriptional regulator involved in sulfur homeostasis with homologs in diverse microorganisms
To Be Published
|
|
4GWO
 
 | |
1UTB
 
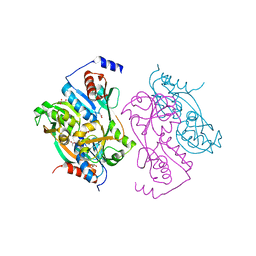 | | DntR from Burkholderia sp. strain DNT | | Descriptor: | ACETATE ION, GLYCEROL, LYSR-TYPE REGULATORY PROTEIN | | Authors: | Smirnova, I.A, Dian, C, Leonard, G.A, McSweeney, S, Birse, D, Brzezinski, P. | | Deposit date: | 2003-12-05 | | Release date: | 2004-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Development of a Bacterial Biosensor for Nitrotoluenes: The Crystal Structure of the Transcriptional Regulator Dntr
J.Mol.Biol., 340, 2004
|
|
6XTU
 
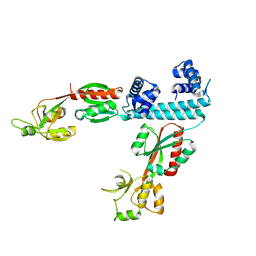 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM | | Descriptor: | Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
6XTV
 
 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM WITH BOUND EFFECTOR ARG | | Descriptor: | ARGININE, Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
8J4N
 
 | |
8J4O
 
 | |
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | Descriptor: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1IXC
 
 | | Crystal structure of CbnR, a LysR family transcriptional regulator | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-06-18 | | Release date: | 2003-06-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
1IZ1
 
 | | CRYSTAL STRUCTURE OF CBNR, A LYSR FAMILY TRANSCRIPTIONAL REGULATOR | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-09-18 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
2UYE
 
 | | Double mutant Y110S,F111V DntR from Burkholderia sp. strain DNT in complex with thiocyanate | | Descriptor: | GLYCEROL, REGULATORY PROTEIN, THIOCYANATE ION | | Authors: | Lonneborg, R, Smirova, I, Dian, C, leonard, G.A, McSweeney, S, Brzezinski, P. | | Deposit date: | 2007-04-04 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In Vivo and in Vitro Investigation of Transcriptional Regulation by Dntr.
J.Mol.Biol., 372, 2007
|
|
