3SJA
 
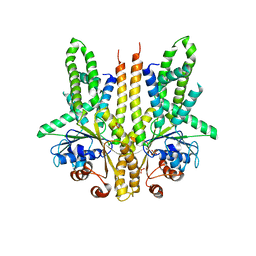 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3B2E
 
 | |
3ZS8
 
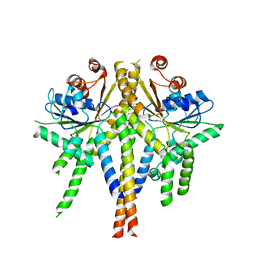 | | S. cerevisiae Get3 complexed with a cytosolic Get1 fragment | | Descriptor: | ATPASE GET3, GOLGI TO ER TRAFFIC PROTEIN 1, ZINC ION | | Authors: | Mariappan, M, Mateja, A, Dobosz, M, Bove, E, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2011-06-24 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Mechanism of Membrane-Associated Steps in Tail-Anchored Protein Insertion.
Nature, 477, 2011
|
|
3SJC
 
 | |
3SJB
 
 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3VLC
 
 | |
