2O3E
 
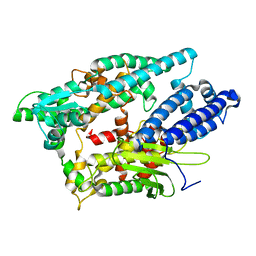 | |
1I1I
 
 | | NEUROLYSIN (ENDOPEPTIDASE 24.16) CRYSTAL STRUCTURE | | Descriptor: | NEUROLYSIN, ZINC ION | | Authors: | Brown, C.K, Madauss, K, Lian, W, Tolbert, W.D, Beck, M.R, Rodgers, D.W. | | Deposit date: | 2001-02-01 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of neurolysin reveals a deep channel that limits substrate access.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4FXY
 
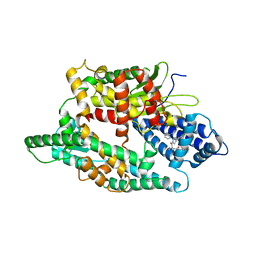 | | Crystal structure of rat neurolysin with bound pyrazolidin inhibitor | | Descriptor: | 1-{(2S)-1-[(3R)-3-(2-chlorophenyl)-2-(2-fluorophenyl)pyrazolidin-1-yl]-1-oxopropan-2-yl}-3-[(1R,3S,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-2-yl]urea, Neurolysin, mitochondrial, ... | | Authors: | Rodgers, D.W, Hines, C.S. | | Deposit date: | 2012-07-03 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric inhibition of the neuropeptidase neurolysin.
J.Biol.Chem., 289, 2014
|
|
