2AL1
 
 | | Crystal Structure Analysis of Enolase Mg Subunit Complex at pH 8.0 | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sims, P.A, Menefee, A.L, Larsen, T.M, Mansoorabadi, S.O, Reed, G.H. | | Deposit date: | 2005-08-04 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and catalytic properties of an engineered heterodimer of enolase composed of one active and one inactive subunit
J.Mol.Biol., 355, 2006
|
|
2XH0
 
 | |
2XH4
 
 | |
2XH7
 
 | |
1ONE
 
 | | YEAST ENOLASE COMPLEXED WITH AN EQUILIBRIUM MIXTURE OF 2'-PHOSPHOGLYCEATE AND PHOSPHOENOLPYRUVATE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, ENOLASE, MAGNESIUM ION, ... | | Authors: | Larsen, T.M, Wedekind, J.E, Rayment, I, Reed, G.H. | | Deposit date: | 1995-12-05 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A carboxylate oxygen of the substrate bridges the magnesium ions at the active site of enolase: structure of the yeast enzyme complexed with the equilibrium mixture of 2-phosphoglycerate and phosphoenolpyruvate at 1.8 A resolution.
Biochemistry, 35, 1996
|
|
2XH2
 
 | |
2XGZ
 
 | |
1P43
 
 | | REVERSE PROTONATION IS THE KEY TO GENERAL ACID-BASE CATALYSIS IN ENOLASE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Enolase 1, MAGNESIUM ION | | Authors: | Sims, P.A, Larsen, T.M, Poyner, R.R, Cleland, W.W, Reed, G.H. | | Deposit date: | 2003-04-21 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reverse protonation is the key to general acid-base catalysis in enolase
Biochemistry, 42, 2003
|
|
2AL2
 
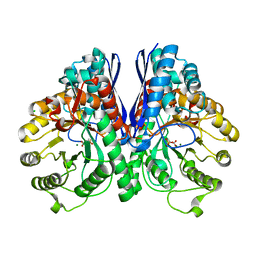 | | Crystal Structure Analysis of Enolase Mg Subunit Complex at pH 8.0 | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sims, P.A, Menefee, A.L, Larsen, T.M, Mansoorabadi, S.O, Reed, G.H. | | Deposit date: | 2005-08-04 | | Release date: | 2006-01-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and catalytic properties of an engineered heterodimer of enolase composed of one active and one inactive subunit
J.Mol.Biol., 355, 2006
|
|
1EBH
 
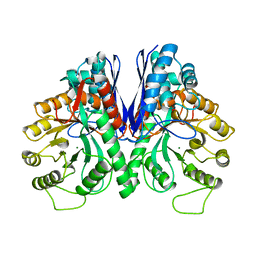 | |
4ENL
 
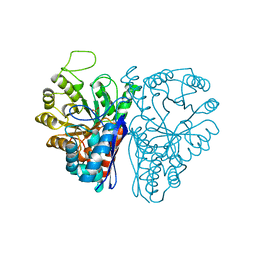 | |
2ONE
 
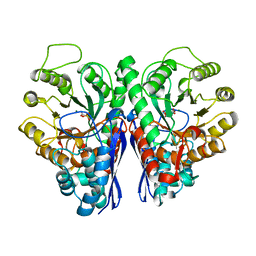 | |
1P48
 
 | | REVERSE PROTONATION IS THE KEY TO GENERAL ACID-BASE CATALYSIS IN ENOLASE | | Descriptor: | Enolase 1, MAGNESIUM ION, PHOSPHOENOLPYRUVATE | | Authors: | Sims, P.A, Larsen, T.M, Poyner, R.R, Cleland, W.W, Reed, G.H. | | Deposit date: | 2003-04-21 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reverse protonation is the key to general acid-base catalysis in enolase
Biochemistry, 42, 2003
|
|
1L8P
 
 | | Mg-phosphonoacetohydroxamate complex of S39A yeast enolase 1 | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETOHYDROXAMIC ACID, enolase 1 | | Authors: | Poyner, R.R, Larsen, T.M, Wong, S.W, Reed, G.H. | | Deposit date: | 2002-03-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase.
Arch.Biochem.Biophys., 401, 2002
|
|
1EBG
 
 | |
7ENL
 
 | |
5ENL
 
 | |
6ENL
 
 | |
3ENL
 
 | |
1ELS
 
 | | CATALYTIC METAL ION BINDING IN ENOLASE: THE CRYSTAL STRUCTURE OF ENOLASE-MN2+-PHOSPHONOACETOHYDROXAMATE COMPLEX AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | ENOLASE, MANGANESE (II) ION, PHOSPHONOACETOHYDROXAMIC ACID | | Authors: | Zhang, E, Hatada, M, Brewer, J.M, Lebioda, L. | | Deposit date: | 1994-04-05 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic metal ion binding in enolase: the crystal structure of an enolase-Mn2+-phosphonoacetohydroxamate complex at 2.4-A resolution.
Biochemistry, 33, 1994
|
|
1NEL
 
 | | FLUORIDE INHIBITION OF YEAST ENOLASE: CRYSTAL STRUCTURE OF THE ENOLASE-MG2+-F--PI COMPLEX AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ENOLASE, FLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Lebioda, L, Zhang, E, Lewinski, K, Brewer, M.J. | | Deposit date: | 1993-08-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fluoride inhibition of yeast enolase: crystal structure of the enolase-Mg(2+)-F(-)-Pi complex at 2.6 A resolution.
Proteins, 16, 1993
|
|
