4A1K
 
 | | Ykud L,D-transpeptidase | | Descriptor: | PUTATIVE L, D-TRANSPEPTIDASE YKUD, SULFATE ION | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Three New Crystal Forms of the Ykud L,D-Transpeptidase from B. Subtilis.
To be Published
|
|
4A1I
 
 | | ykud from B.subtilis | | Descriptor: | CADMIUM ION, CHLORIDE ION, PUTATIVE L, ... | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of Three New Crystal Forms of the Ykud L,D-Transpeptidase from B. Subtilis.
To be Published
|
|
1Y7M
 
 | | Crystal Structure of the B. subtilis YkuD protein at 2 A resolution | | Descriptor: | CADMIUM ION, SULFATE ION, hypothetical protein BSU14040 | | Authors: | Bielnicki, J.A, Devedjiev, Y, Derewenda, U, Dauter, Z, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-09 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | B. subtilis ykuD protein at 2.0 A resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes.
Proteins, 62, 2006
|
|
4A1J
 
 | | Ykud L,D-transpeptidase from B.subtilis | | Descriptor: | PUTATIVE L, D-TRANSPEPTIDASE YKUD, SULFATE ION | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Ykud
To be Published
|
|
2MTZ
 
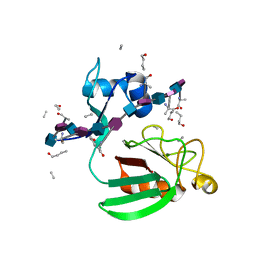 | | Haddock model of Bacillus subtilis L,D-transpeptidase in complex with a peptidoglycan hexamuropeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Putative L,D-transpeptidase YkuD, intact bacterial peptidoglycan | | Authors: | Schanda, P, Triboulet, S, Laguri, C, Bougault, C, Ayala, I, Callon, M, Arthur, M, Simorre, J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of a cell-wall cross-linking enzyme in complex with an intact bacterial peptidoglycan.
J.Am.Chem.Soc., 136, 2014
|
|
3ZQD
 
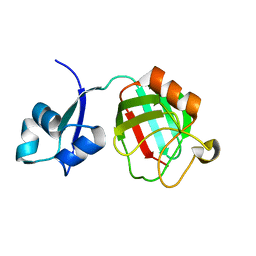 | | B. subtilis L,D-transpeptidase | | Descriptor: | L, D-TRANSPEPTIDASE YKUD | | Authors: | Lecoq, L, Simorre, J.-P, Bougault, C, Arthur, M, Hugonnet, J.-E, Veckerle, C, Pessey, O. | | Deposit date: | 2011-06-09 | | Release date: | 2012-05-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
4A52
 
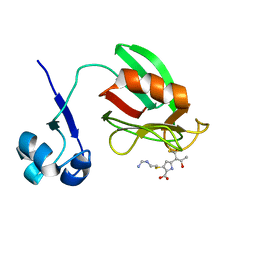 | | NMR structure of the imipenem-acylated L,D-transpeptidase from Bacillus subtilis | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, PUTATIVE L, D-TRANSPEPTIDASE YKUD | | Authors: | Lecoq, L, Simorre, J, Bougault, C, Arthur, M, Hugonnet, J, Veckerle, C, Pessey, O. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-30 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
