3NED
 
 | | mRouge | | Descriptor: | ACETATE ION, PAmCherry1 protein, SODIUM ION | | Authors: | Mayo, S.L, Chica, R.A, Moore, M.M. | | Deposit date: | 2010-06-08 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Generation of longer emission wavelength red fluorescent proteins using computationally designed libraries.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
6MZ3
 
 | | mCherry pH sensitive mutant - M66T (mCherryTYG) | | Descriptor: | PAmCherry1 protein | | Authors: | Haynes, E.P, Tantama, M. | | Deposit date: | 2018-11-03 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.088 Å) | | Cite: | Quantifying Acute Fuel and Respiration Dependent pH Homeostasis in Live Cells Using the mCherryTYG Mutant as a Fluorescence Lifetime Sensor.
Anal.Chem., 91, 2019
|
|
3LF3
 
 | | Crystal Structure of Fast Fluorescent Timer Fast-FT | | Descriptor: | Fast Fluorescent Timer Fast-FT | | Authors: | Pletnev, S, Dauter, Z. | | Deposit date: | 2010-01-15 | | Release date: | 2010-03-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Understanding blue-to-red conversion in monomeric fluorescent timers and hydrolytic degradation of their chromophores
J.Am.Chem.Soc., 132, 2010
|
|
7SAK
 
 | |
8B7G
 
 | | Structure of oxygen-degraded rsCherry | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bui, T.Y.H, Van Meervelt, L. | | Deposit date: | 2022-09-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxygen-induced chromophore degradation in the photoswitchable red fluorescent protein rsCherry.
Int.J.Biol.Macromol., 239, 2023
|
|
3KCS
 
 | | Crystal structure of PAmCherry1 in the dark state | | Descriptor: | PAmCherry1 protein | | Authors: | Malashkevich, V.N, Subach, F.V, Zencheck, W.D, Xiao, H, Filonov, G.S, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8B65
 
 | | Structure of rsCherry crystallized in anaerobic conditions | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bui, T.Y.H, Van Meervelt, L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Oxygen-induced chromophore degradation in the photoswitchable red fluorescent protein rsCherry.
Int.J.Biol.Macromol., 239, 2023
|
|
6YLM
 
 | | mCherry | | Descriptor: | CHLORIDE ION, mCherry | | Authors: | Myskova, J, Rybakova, M, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3KCT
 
 | | CRYSTAL STRUCTURE OF PAmCherry1 in the photoactivated state | | Descriptor: | PAmCherry1 protein | | Authors: | Malashkevich, V.N, Subach, F.V, Zencheck, W.D, Xiao, H, Filonov, G.S, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3NEZ
 
 | | mRojoA | | Descriptor: | mRojoA | | Authors: | Mayo, S.L, Chica, R.A, Moore, M.M. | | Deposit date: | 2010-06-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Generation of longer emission wavelength red fluorescent proteins using computationally designed libraries.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5H89
 
 | | Crystal structure of mRojoA mutant - T16V - P63Y - W143G - L163V | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
3LF4
 
 | | Crystal Structure of Fluorescent Timer Precursor Blue102 | | Descriptor: | Fluorescent Timer Precursor Blue102 | | Authors: | Pletnev, S, Dauter, Z. | | Deposit date: | 2010-01-15 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | Understanding blue-to-red conversion in monomeric fluorescent timers and hydrolytic degradation of their chromophores
J.Am.Chem.Soc., 132, 2010
|
|
7SAL
 
 | |
8BGL
 
 | | Structure of the dimeric rsCherryRev1.4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PAmCherry1 protein, ... | | Authors: | Bui, T.Y.H, Van Meervelt, L. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An unusual disulfide-linked dimerization in the fluorescent protein rsCherryRev1.4.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7RY2
 
 | | mSandy2 | | Descriptor: | mSandy2 | | Authors: | Chica, R.A, Legault, S, Thompson, M.C. | | Deposit date: | 2021-08-24 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Generation of bright monomeric red fluorescent proteins via computational design of enhanced chromophore packing.
Chem Sci, 13, 2022
|
|
5H87
 
 | | Crystal structure of mRojoA mutant - P63H - W143S | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
7SAJ
 
 | |
8K9T
 
 | | Cryo-EM structure of the products-bound PGAP1(Bst1)-S327A from Chaetonium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8K9R
 
 | | Cryo EM structure of the products-bound PGAP1(Bst1)-H443N from Chaetomium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
6B0B
 
 | | Crystal structure of human APOBEC3H | | Descriptor: | APOBEC3H, MCherry, RNA (5'-R(*UP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Shaban, N.M, Shi, K, Banerjee, S, Harris, R.S, Aihara, H. | | Deposit date: | 2017-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.2800622 Å) | | Cite: | The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNA-Mediated Dimerization Mechanism.
Mol. Cell, 69, 2018
|
|
7WUJ
 
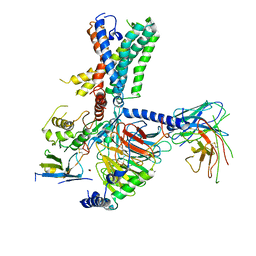 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G4,Uncharacterized protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Guo, S.C, Huang, S.M, He, Q.T, Xiao, P, Sun, J.P, Yu, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
6BBO
 
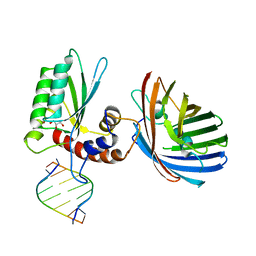 | | Crystal structure of human APOBEC3H/RNA complex | | Descriptor: | APOBEC3H, GLYCEROL, MCherry fluorescent protein, ... | | Authors: | Shaban, N.M, Shi, K, Banerjee, S, Harris, R.S, Aihara, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.428 Å) | | Cite: | The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNA-Mediated Dimerization Mechanism.
Mol. Cell, 69, 2018
|
|
7QPG
 
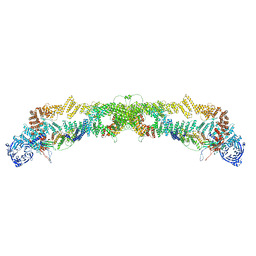 | | Human RZZ kinetochore corona complex. | | Descriptor: | Centromere/kinetochore protein zw10 homolog, Kinetochore-associated protein 1, Protein zwilch homolog | | Authors: | Raisch, T, Ciossani, G, d'Amico, E, Cmetowski, V, Carmignani, S, Maffini, S, Merino, F, Wohlgemuth, S, Vetter, I.R, Raunser, S, Musacchio, A. | | Deposit date: | 2022-01-04 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the RZZ complex and molecular basis of Spindly-driven corona assembly at human kinetochores.
Embo J., 41, 2022
|
|
