4GYJ
 
 | |
4GVH
 
 | |
4GVG
 
 | |
4GVF
 
 | |
4GYK
 
 | |
4GVI
 
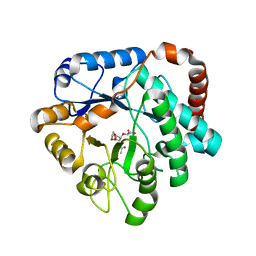 | | Crystal structure of mutant (D248N) Salmonella typhimurium family 3 glycoside hydrolase (NagZ) in complex with GlcNAc-1,6-anhMurNAc | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bacik, J.P, Mark, B.L. | | Deposit date: | 2012-08-30 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Active Site Plasticity within the Glycoside Hydrolase NagZ Underlies a Dynamic Mechanism of Substrate Distortion.
Chem.Biol., 19, 2012
|
|
