1SSN
 
 | | STAPHYLOKINASE, SAKSTAR VARIANT, NMR, 20 STRUCTURES | | Descriptor: | STAPHYLOKINASE | | Authors: | Ohlenschlager, O, Ramachandran, R, Guhrs, K.H, Schlott, B, Brown, L.R. | | Deposit date: | 1998-06-07 | | Release date: | 1998-12-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the plasminogen-activator protein staphylokinase.
Biochemistry, 37, 1998
|
|
1SSO
 
 | | SOLUTION STRUCTURE AND DNA-BINDING PROPERTIES OF A THERMOSTABLE PROTEIN FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS | | Descriptor: | SSO7D | | Authors: | Baumann, H, Knapp, S, Lundback, T, Ladenstein, R, Hard, T. | | Deposit date: | 1995-03-31 | | Release date: | 1995-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus.
Nat.Struct.Biol., 1, 1994
|
|
1SSP
 
 | | WILD-TYPE URACIL-DNA GLYCOSYLASE BOUND TO URACIL-CONTAINING DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*(D1P)P*AP*TP*CP*TP*T)-3', URACIL, ... | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
1SSQ
 
 | | Serine Acetyltransferase- Complex with Cysteine | | Descriptor: | CYSTEINE, MAGNESIUM ION, Serine acetyltransferase | | Authors: | Olsen, L.R, Huang, B, Vetting, M.W, Roderick, S.L. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Serine Acetyltransferase in Complexes with CoA and its Cysteine Feedback Inhibitor
Biochemistry, 43, 2004
|
|
1SST
 
 | | Serine Acetyltransferase- Complex with CoA | | Descriptor: | COENZYME A, Serine acetyltransferase | | Authors: | Olsen, L.R, Huang, B, Vetting, M.W, Roderick, S.L. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Serine Acetyltransferase in Complexes with CoA and its Cysteine Feedback Inhibitor
Biochemistry, 43, 2004
|
|
1SSU
 
 | | Structural and biochemical evidence for disulfide bond heterogeneity in active forms of the somatomedin B domain of human vitronectin | | Descriptor: | Vitronectin | | Authors: | Kamikubo, Y, De Guzman, R, Kroon, G, Curriden, S, Neels, J.G, Churchill, M.J, Dawson, P, Oldziej, S, Jagielska, A, Scheraga, H.A, Loskutoff, D.J, Dyson, H.J. | | Deposit date: | 2004-03-24 | | Release date: | 2004-07-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Disulfide bonding arrangements in active forms of the somatomedin B domain of human vitronectin.
Biochemistry, 43, 2004
|
|
1SSV
 
 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
1SSW
 
 | | Crystal structure of phage T4 lysozyme mutant Y24A/Y25A/T26A/I27A/C54T/C97A | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme | | Authors: | He, M.M, Baase, W.A, Xiao, H, Heinz, D.W, Matthews, B.W. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation
Protein Sci., 13, 2004
|
|
1SSX
 
 | |
1SSY
 
 | | Crystal structure of phage T4 lysozyme mutant G28A/I29A/G30A/C54T/C97A | | Descriptor: | Lysozyme | | Authors: | He, M.M, Baase, W.A, Xiao, H, Heinz, D.W, Matthews, B.W. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation
Protein Sci., 13, 2004
|
|
1SSZ
 
 | | Conformational Mapping of Mini-B: An N-terminal/C-terminal Construct of Surfactant Protein B Using 13C-Enhanced Fourier Transform Infrared (FTIR) Spectroscopy | | Descriptor: | Pulmonary surfactant-associated protein B | | Authors: | Waring, A.J, Walther, F.J, Gordon, L.M, Hernandez-Juviel, J.M, Hong, T, Sherman, M.A, Alonso, C, Alig, T, Braun, A, Bacon, D, Zasadzinski, J.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-30 | | Method: | INFRARED SPECTROSCOPY | | Cite: | The role of charged amphipathic helices in the structure and function of surfactant protein B.
J.Pept.Res., 66, 2005
|
|
1ST0
 
 | | Structure of DcpS bound to m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, YTTRIUM (III) ION, mRNA decapping enzyme | | Authors: | Gu, M, Fabrega, C, Liu, S.W, Liu, H, Kiledjian, M, Lima, C.D. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity
Mol.Cell, 14, 2004
|
|
1ST2
 
 | |
1ST3
 
 | |
1ST4
 
 | | Structure of DcpS bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, YTTRIUM (III) ION, mRNA decapping enzyme | | Authors: | Gu, M, Fabrega, C, Liu, S.W, Liu, H, Kiledjian, M, Lima, C.D. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity
Mol.Cell, 14, 2004
|
|
1ST6
 
 | |
1ST7
 
 | | Solution structure of Acyl Coenzyme A Binding Protein from yeast | | Descriptor: | Acyl-CoA-binding protein | | Authors: | Teilum, K, Thormann, T, Caterer, N.R, Poulsen, H.I, Jensen, P.H, Knudsen, J, Kragelund, B.B, Poulsen, F.M. | | Deposit date: | 2004-03-25 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Different secondary structure elements as scaffolds for protein folding transition states of two homologous four-helix bundles
Proteins, 59, 2005
|
|
1ST8
 
 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Verhaest, M, Van den Ende, W, De Ranter, C.J, Van Laere, A, Rabijns, A. | | Deposit date: | 2004-03-25 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus.
Plant J., 41, 2005
|
|
1ST9
 
 | | Crystal Structure of a Soluble Domain of ResA in the Oxidised Form | | Descriptor: | 1,2-ETHANEDIOL, Thiol-disulfide oxidoreductase resA | | Authors: | Crow, A, Acheson, R.M, Le Brun, N.E, Oubrie, A. | | Deposit date: | 2004-03-25 | | Release date: | 2004-05-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Redox-coupled Protein Substrate Selection by the Cytochrome c Biosynthesis Protein ResA.
J.Biol.Chem., 279, 2004
|
|
1STA
 
 | | ACCOMMODATION OF INSERTION MUTATIONS ON THE SURFACE AND IN THE INTERIOR OF STAPHYLOCOCCAL NUCLEASE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Keefe, L.J, Quirk, S, Gittis, A, Lattman, E.E. | | Deposit date: | 1994-01-17 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of insertion mutations on the surface and in the interior of staphylococcal nuclease.
Protein Sci., 3, 1994
|
|
1STB
 
 | | ACCOMMODATION OF INSERTION MUTATIONS ON THE SURFACE AND IN THE INTERIOR OF STAPHYLOCOCCAL NUCLEASE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Quirk, S, Gittis, A, Keefe, L.J, Lattman, E.E. | | Deposit date: | 1994-01-17 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Accommodation of insertion mutations on the surface and in the interior of staphylococcal nuclease.
Protein Sci., 3, 1994
|
|
1STC
 
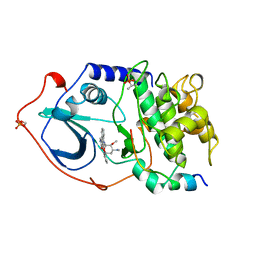 | | CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH STAUROSPORINE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR, STAUROSPORINE | | Authors: | Prade, L, Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1997-10-10 | | Release date: | 1998-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staurosporine-induced conformational changes of cAMP-dependent protein kinase catalytic subunit explain inhibitory potential.
Structure, 5, 1997
|
|
1STD
 
 | |
1STE
 
 | |
1STF
 
 | |
