1L98
 
 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | Deposit date: | 1992-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
1L99
 
 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | Deposit date: | 1992-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
1L9A
 
 | | CRYSTAL STRUCTURE OF SRP19 IN COMPLEX WITH THE S DOMAIN OF SIGNAL RECOGNITION PARTICLE RNA | | Descriptor: | MAGNESIUM ION, METHYL MERCURY ION, SIGNAL RECOGNITION PARTICLE 19 KDA PROTEIN, ... | | Authors: | Oubridge, C, Kuglstatter, A, Jovine, L, Nagai, K. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle.
Mol.Cell, 9, 2002
|
|
1L9B
 
 | | X-Ray Structure of the Cytochrome-c(2)-Photosynthetic Reaction Center Electron Transfer Complex from Rhodobacter sphaeroides in Type II Co-Crystals | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CHLORIDE ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Okamura, M.Y, Yeh, A.P, Rees, D.C, Feher, G. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure determination of the cytochrome c2: reaction center electron transfer complex from Rhodobacter sphaeroides.
J.Mol.Biol., 319, 2002
|
|
1L9C
 
 | | Role of Histidine 269 in Catalysis by Monomeric Sarcosine Oxidase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric Sarcosine Oxidase, ... | | Authors: | Zhao, G, Song, H, Chen, Z.-w, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Monomeric sarcosine oxidase: role of histidine 269 in catalysis.
Biochemistry, 41, 2002
|
|
1L9D
 
 | | Role of Histidine 269 in Catalysis by Monomeric Sarcosine Oxidase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase, ... | | Authors: | Zhao, G, Song, H, Chen, Z.-w, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-08-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Monomeric sarcosine oxidase: role of histidine 269 in catalysis.
Biochemistry, 41, 2002
|
|
1L9E
 
 | | Role of Histidine 269 in Catalysis by Monomeric Sarcosine Oxidase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Zhao, G, Song, H, Chen, Z.-w, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Monomeric sarcosine oxidase: role of histidine 269 in catalysis.
Biochemistry, 41, 2002
|
|
1L9G
 
 | | CRYSTAL STRUCTURE OF URACIL-DNA GLYCOSYLASE FROM T. MARITIMA | | Descriptor: | Conserved hypothetical protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Rajashankar, K.R, Dodatko, T, Thirumuruhan, R.A, Sandigursky, M, Bresnik, A, Chance, M.R, Franklin, W.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-22 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of uracil-DNA glycosylase from T. Maritima
To be Published
|
|
1L9H
 
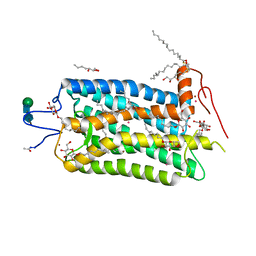 | | Crystal structure of bovine rhodopsin at 2.6 angstroms RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Okada, T, Fujiyoshi, Y, Silow, M, Navarro, J, Landau, E.M, Shichida, Y. | | Deposit date: | 2002-03-23 | | Release date: | 2002-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1L9J
 
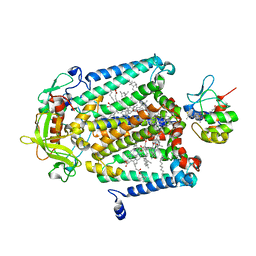 | | X-Ray Structure of the Cytochrome-c(2)-Photosynthetic Reaction Center Electron Transfer Complex from Rhodobacter sphaeroides in Type I Co-Crystals | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CHLORIDE ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Okamura, M.Y, Yeh, A.P, Rees, D.C, Feher, G. | | Deposit date: | 2002-03-24 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | X-ray structure determination of the cytochrome c2: reaction center electron transfer complex from Rhodobacter sphaeroides.
J.Mol.Biol., 319, 2002
|
|
1L9K
 
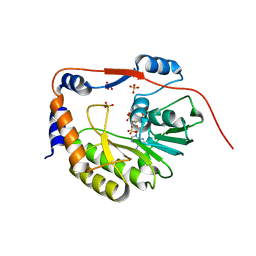 | | dengue methyltransferase | | Descriptor: | RNA-DIRECTED RNA POLYMERASE, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Egloff, M.P, Benarroch, D, Selisko, B, Romette, J.L, Canard, B. | | Deposit date: | 2002-03-25 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An RNA cap (nucleoside-2'-O-) methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization
Embo J., 21, 2002
|
|
1L9L
 
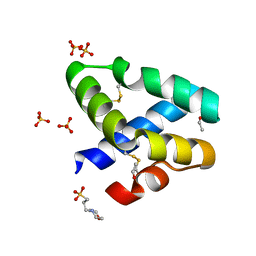 | | GRANULYSIN FROM HUMAN CYTOLYTIC T LYMPHOCYTES | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ETHANOL, Granulysin, ... | | Authors: | Anderson, D.H, Sawaya, M.R, Cascio, D, Ernst, W, Krensky, A, Modlin, R, Eisenberg, D. | | Deposit date: | 2002-03-25 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Granulysin Crystal Structure and a Structure-Derived Lytic Mechanism
J.Mol.Biol., 325, 2002
|
|
1L9M
 
 | |
1L9N
 
 | |
1L9O
 
 | |
1L9P
 
 | |
1L9Q
 
 | |
1L9R
 
 | |
1L9S
 
 | |
1L9T
 
 | |
1L9U
 
 | | THERMUS AQUATICUS RNA POLYMERASE HOLOENZYME AT 4 A RESOLUTION | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Darst, S.A. | | Deposit date: | 2002-03-26 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution.
Science, 296, 2002
|
|
1L9V
 
 | | Non Structural protein encoded by gene segment 8 of rotavirus (NSP2), an NTPase, ssRNA binding and nucleic acid helix-destabilizing protein | | Descriptor: | Rotavirus-NSP2 | | Authors: | Jayaram, H, Taraporewala, Z, Patton, J.T, Prasad, B.V. | | Deposit date: | 2002-03-26 | | Release date: | 2002-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold.
Nature, 417, 2002
|
|
1L9W
 
 | | CRYSTAL STRUCTURE OF 3-DEHYDROQUINASE FROM SALMONELLA TYPHI COMPLEXED WITH REACTION PRODUCT | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, 3-dehydroquinate dehydratase aroD | | Authors: | Lee, W.H, Perles, L.A, Nagem, R.A.P, Shrive, A.K, Hawkins, A, Sawyer, L, Polikarpov, I. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of different crystal forms of 3-dehydroquinase from Salmonella typhi and its implication for the enzyme activity.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1L9X
 
 | | Structure of gamma-Glutamyl Hydrolase | | Descriptor: | BETA-MERCAPTOETHANOL, gamma-glutamyl hydrolase | | Authors: | Li, H, Ryan, T.J, Chave, K.J, Van Roey, P. | | Deposit date: | 2002-03-26 | | Release date: | 2002-04-10 | | Last modified: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structure of human gamma -glutamyl hydrolase. A class I glatamine amidotransferase adapted for a complex substate.
J.Biol.Chem., 277, 2002
|
|
1L9Y
 
 | | FEZ-1-Y228A, A Mutant of the Metallo-beta-lactamase from Legionella gormanii | | Descriptor: | CHLORIDE ION, FEZ-1 b-lactamase, GLYCEROL, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Galleni, M, Dideberg, O. | | Deposit date: | 2002-03-27 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Three-dimensional Structure of FEZ-1, a Monomeric Subclass B3 Metallo-beta-lactamase
from Fluoribacter gormanii, in Native Form and in Complex with -Captopril
J.Mol.Biol., 325, 2003
|
|
