2JA6
 
 | | CPD lesion containing RNA Polymerase II elongation complex B | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TTP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | CPD damage recognition by transcribing RNA polymerase II.
Science, 315, 2007
|
|
2JA5
 
 | | CPD lesion containing RNA Polymerase II elongation complex A | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TTP*TP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II
Science, 315, 2007
|
|
4WRB
 
 | | Macrophage Migration Inhibitory Factor in complex with a biaryltriazole inhibitor (3b-190) | | Descriptor: | 4-{4-[6-(2-methoxyethoxy)quinolin-2-yl]-1H-1,2,3-triazol-1-yl}phenol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Robertson, M.J, Baxter, R.H.G, Jorgensen, W.L. | | Deposit date: | 2014-10-23 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Design, synthesis, and protein crystallography of biaryltriazoles as potent tautomerase inhibitors of macrophage migration inhibitory factor.
J.Am.Chem.Soc., 137, 2015
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | Descriptor: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
7BKV
 
 | | Endothiapepsin structure obtained at 100K with fragment AC39729 bound | | Descriptor: | 5-fluoranylpyridin-2-amine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Engilberge, S, Huang, C.-Y, Smith, K.M.L, Eris, D, Marsh, M, Wang, M, Wojdyla, J.A. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Endothiapepsin structure obtained at 100K with fragment AC39729 bound
To Be Published
|
|
4XIS
 
 | |
7CJO
 
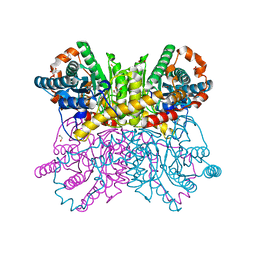 | | Crystal structure of metal-bound state of glucose isomerase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Xylose isomerase | | Authors: | Nam, K.H. | | Deposit date: | 2020-07-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the metal-free state of glucose isomerase reveals its minimal open configuration for metal binding.
Biochem.Biophys.Res.Commun., 547, 2021
|
|
7CJP
 
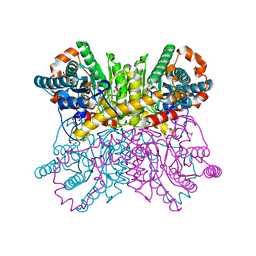 | | Crystal structure of metal-free state of glucose isomerase | | Descriptor: | 1,2-ETHANEDIOL, Xylose isomerase | | Authors: | Nam, K.H. | | Deposit date: | 2020-07-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the metal-free state of glucose isomerase reveals its minimal open configuration for metal binding.
Biochem.Biophys.Res.Commun., 547, 2021
|
|
9LNP
 
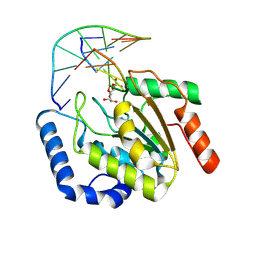 | | the hUNG bound to DNA product embedding uridine ribonucleotide | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*T*(RP5)P*AP*TP*CP*TP*T)-3'), ... | | Authors: | Liu, Y, Zhou, C, Zhan, X, Fan, C. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Uridine Embedded within DNA is Repaired by Uracil DNA Glycosylase via a Mechanism Distinct from That of Ribonuclease H2.
J.Am.Chem.Soc., 147, 2025
|
|
5QR1
 
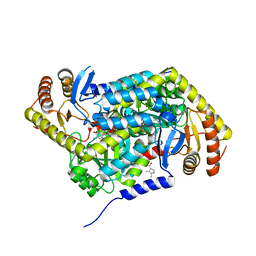 | | PanDDA analysis group deposition -- Crystal Structure of human ALAS2A in complex with Z396380540 | | Descriptor: | 5-aminolevulinate synthase, erythroid-specific, mitochondrial, ... | | Authors: | Bezerra, G.A, Foster, W, Bailey, H, Shrestha, L, Krojer, T, Talon, R, Brandao-Neto, J, Douangamath, A, Nicola, B.B, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Brennan, P.E, Yue, W.W. | | Deposit date: | 2019-05-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6A0H
 
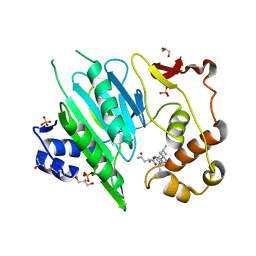 | |
6A0F
 
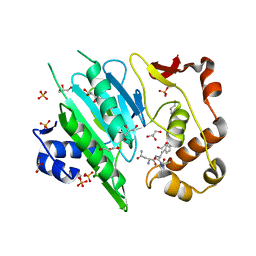 | |
8QPL
 
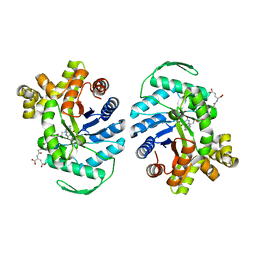 | | F420-Dependent Methylene-Tetrahydromethanopterin Reductase with F420 from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase, COENZYME F420 | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
6YOY
 
 | |
8F78
 
 | | Compound 1 bound to procaspase-6 | | Descriptor: | 5-fluoro-2-({[3-(pyrimidin-2-yl)pyridin-2-yl]amino}methyl)phenol, CHLORIDE ION, Procaspase-6 | | Authors: | Fan, P, Zhao, Y, Renslo, A.R, Arkin, M.R. | | Deposit date: | 2022-11-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Study of Heteroarene Stacking Using a Congeneric Set of Molecular Glues for Procaspase-6.
J.Med.Chem., 66, 2023
|
|
8FHM
 
 | | RNase A-Uridine 5'-Hexaphosphate (RNaseA.p6U) | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy{[(R)-hydroxy{[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]oxy}phosphoryl]oxy}phosphoryl]uridine, Ribonuclease pancreatic | | Authors: | Park, G, Cummins, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION SCATTERING (1.79 Å), X-RAY DIFFRACTION | | Cite: | Pentaphosphorylation via the Anhydride of Dihydrogen Pentametaphosphate: Access to Nucleoside Hexa- and Heptaphosphates and Study of Their Interaction with Ribonuclease A.
Acs Cent.Sci., 10, 2024
|
|
8FOQ
 
 | |
7OSQ
 
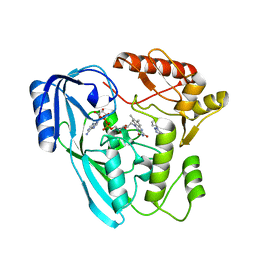 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 5-methyl-1-phenyl-1,2,3-triazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Mayol-Llinas, J, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
4R1E
 
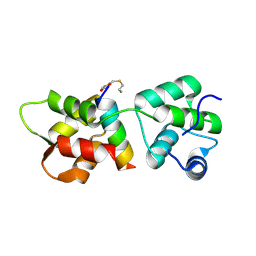 | | Crystal Structure of MTIP from Plasmodium falciparum in complex with a peptide-fragment chimera | | Descriptor: | 5-{[(2-aminoethyl)sulfanyl]methyl}furan-2-carbaldehyde, Myosin A tail domain interacting protein, Myosin-A | | Authors: | Douse, C.H, Vrielink, N, Cota, E, Tate, E.W. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Targeting a Dynamic Protein-Protein Interaction: Fragment Screening against the Malaria Myosin A Motor Complex.
Chemmedchem, 10, 2015
|
|
2JA7
 
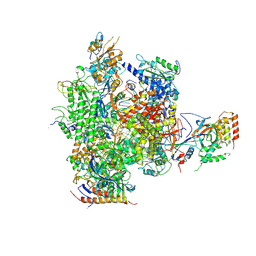 | | CPD lesion containing RNA Polymerase II elongation complex C | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TP*TTP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II.
Science, 315, 2007
|
|
2KH3
 
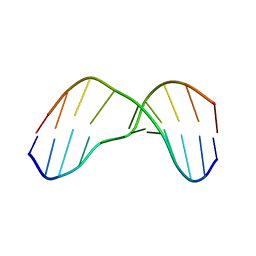 | | NMR Structure of Aflatoxin Formamidopyrimidine alpha-anomer in duplex DNA | | Descriptor: | 5'-D(*CP*TP*AP*TP*(FAG)P*AP*TP*TP*CP*A)-3', 5'-D(*TP*GP*AP*AP*TP*CP*AP*TP*AP*G)-3' | | Authors: | Brown, K.L. | | Deposit date: | 2009-03-24 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural perturbations induced by the alpha-anomer of the aflatoxin B(1) formamidopyrimidine adduct in duplex and single-strand DNA
J.Am.Chem.Soc., 131, 2009
|
|
5RVQ
 
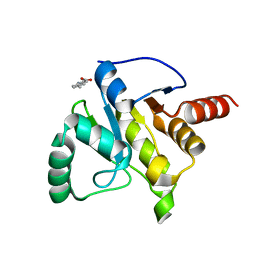 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000002508153 | | Descriptor: | 5-methyl-1H-indole-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7PVM
 
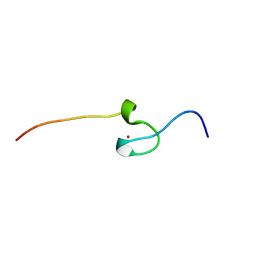 | |
5RTQ
 
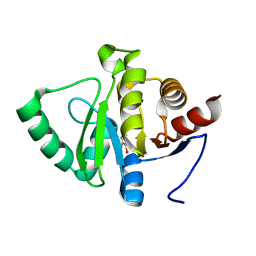 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000019015078 | | Descriptor: | 5-bromo-6-methylpyridin-2-amine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVS
 
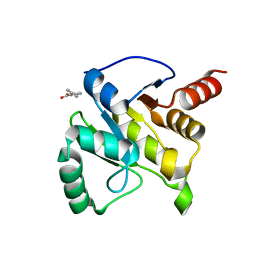 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000159004 | | Descriptor: | 5-phenylpyridine-3-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
