1S7D
 
 | |
1SAC
 
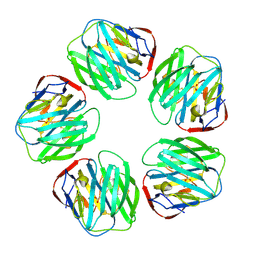 | | THE STRUCTURE OF PENTAMERIC HUMAN SERUM AMYLOID P COMPONENT | | Descriptor: | ACETIC ACID, CALCIUM ION, SERUM AMYLOID P COMPONENT | | Authors: | White, H.E, Emsley, J, O'Hara, B.P, Oliva, G, Srinivasan, N, Tickle, I.J, Blundell, T.L, Pepys, M.B, Wood, S.P. | | Deposit date: | 1994-01-27 | | Release date: | 1994-05-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of pentameric human serum amyloid P component.
Nature, 367, 1994
|
|
1SEU
 
 | | Human DNA Topoisomerase I (70 Kda) In Complex With The Indolocarbazole SA315F and Covalent Complex With A 22 Base Pair DNA Duplex | | Descriptor: | 2,10-DIHYDROXY-12-(BETA-D-GLUCOPYRANOSYL)-6,7,12,13-TETRAHYDROINDOLO[2,3-A]PYRROLO[3,4-C]CARBAZOLE-5,7-DIONE, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', ... | | Authors: | Staker, B.L, Feese, M.D, Cushman, M, Pommier, Y, Zembower, D, Stewart, L, Burgin, A.B. | | Deposit date: | 2004-02-18 | | Release date: | 2005-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex
J.Med.Chem., 48, 2005
|
|
1RZL
 
 | | RICE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, NONSPECIFIC LIPID TRANSFER PROTEIN, SULFATE ION | | Authors: | Lee, J.Y, Min, K.S, Cha, H, Shin, D.H, Hwang, K.Y, Suh, S.W. | | Deposit date: | 1997-10-09 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rice non-specific lipid transfer protein: the 1.6 A crystal structure in the unliganded state reveals a small hydrophobic cavity.
J.Mol.Biol., 276, 1998
|
|
1RYE
 
 | | Crystal Structure of the Shifted Form of the Glucose-Fructose Oxidoreductase from Zymomonas mobilis | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kim, Y, Arora, M, Straza, M, Donnelly, M, Joachimiak, A. | | Deposit date: | 2003-12-22 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Shifted Form of the Glucose-Fructose Oxidoreductase from Zymomonas mobilis
To be Published
|
|
1S0F
 
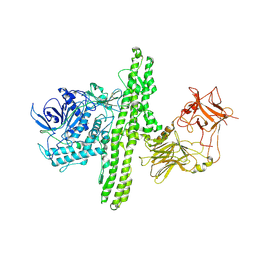 | | Crystal structure of botulinum neurotoxin type B at pH 7.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1S1J
 
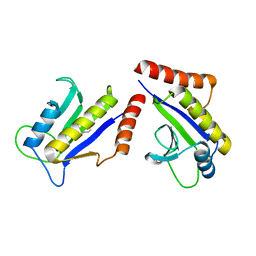 | | Crystal Structure of ZipA in complex with indoloquinolizin inhibitor 1 | | Descriptor: | (12bS)-1,2,3,4,12,12b-hexahydroindolo[2,3-a]quinolizin-7(6H)-one, Cell division protein zipA | | Authors: | Jenning, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenney, C.H, Moghazeh, S.L, Peterson, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | Deposit date: | 2004-01-06 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1S6H
 
 | | PORCINE TRYPSIN COMPLEXED WITH GUANIDINE-3-PROPANOL INHIBITOR | | Descriptor: | 4-(HYDROXYMETHYL)BENZAMIDINE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, Derose, E.F, London, R.E. | | Deposit date: | 2004-01-23 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1S2R
 
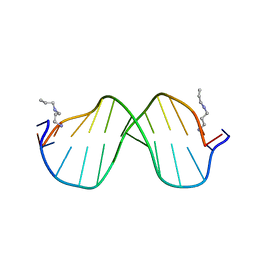 | | A High Resolution Crystal Structure of [d(CGCAAATTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3', SPERMINE | | Authors: | Woods, K.K, Maehigashi, T, Howerton, S.B, Tannenbaum, S, Williams, L.D. | | Deposit date: | 2004-01-09 | | Release date: | 2005-01-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High-resolution structure of an extended A-tract: [d(CGCAAATTTGCG)]2.
J.Am.Chem.Soc., 126, 2004
|
|
1S3N
 
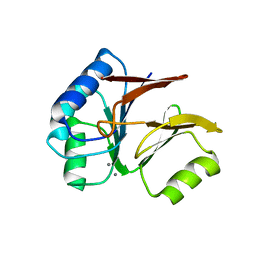 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, MANGANESE (II) ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
1S3Z
 
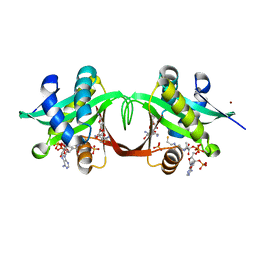 | | Aminoglycoside N-Acetyltransferase AAC(6')-Iy in Complex with CoA and Ribostamycin | | Descriptor: | COENZYME A, NICKEL (II) ION, RIBOSTAMYCIN, ... | | Authors: | Vetting, M.W, Magnet, S, Nieves, E, Roderick, S.L, Blachard, J.S. | | Deposit date: | 2004-01-14 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones
Chem.Biol., 11, 2004
|
|
1SBW
 
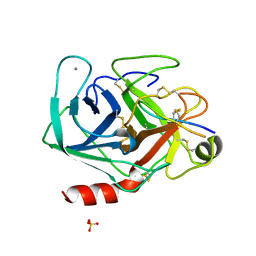 | | CRYSTAL STRUCTURE OF MUNG BEAN INHIBITOR LYSINE ACTIVE FRAGMENT COMPLEX WITH BOVINE BETA-TRYPSIN AT 1.8A RESOLUTION | | Descriptor: | CALCIUM ION, PROTEIN (BETA-TRYPSIN), PROTEIN (MUNG BEAN INHIBITOR LYSIN ACTIVE FRAGMENT), ... | | Authors: | Huang, Q, Zhu, Y, Chi, C, Tang, Y. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mung bean inhibitor lysine active fragment complex with bovine beta-trypsin at 1.8A resolution.
J.Biomol.Struct.Dyn., 16, 1999
|
|
1SHG
 
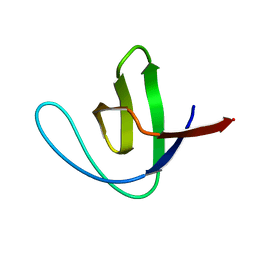 | | CRYSTAL STRUCTURE OF A SRC-HOMOLOGY 3 (SH3) DOMAIN | | Descriptor: | ALPHA-SPECTRIN SH3 DOMAIN | | Authors: | Noble, M, Pauptit, R, Musacchio, A, Saraste, M, Wierenga, R.K. | | Deposit date: | 1993-05-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Src-homology 3 (SH3) domain.
Nature, 359, 1992
|
|
1SHR
 
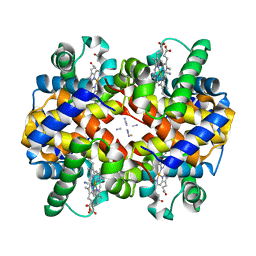 | | Crystal structure of ferrocyanide bound human hemoglobin A2 at 1.88A resolution | | Descriptor: | CYANIDE ION, FE (III) ION, Hemoglobin alpha chain, ... | | Authors: | Sen, U, Dasgupta, J, Choudhury, D, Datta, P, Chakrabarti, A, Chakrabarty, S.B, Chakrabarty, A, Dattagupta, J.K. | | Deposit date: | 2004-02-26 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of HbA2 and HbE and modeling of hemoglobin delta4: interpretation of the thermal stability and the antisickling effect of HbA2 and identification of the ferrocyanide binding site in Hb
Biochemistry, 43, 2004
|
|
1SCE
 
 | |
1RJC
 
 | | Crystal structure of the camelid single domain antibody cAb-Lys2 in complex with hen egg white lysozyme | | Descriptor: | GLYCEROL, Lysozyme C, PHOSPHATE ION, ... | | Authors: | De Genst, E, Silence, K, Ghahroudi, M.A, Decanniere, K, Loris, R, Kinne, J, Wyns, L, Muyldermans, S. | | Deposit date: | 2003-11-19 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Strong in vivo maturation compensates for structurally restricted H3 loops in antibody repertoires.
J.Biol.Chem., 280, 2005
|
|
1RP1
 
 | | DOG PANCREATIC LIPASE RELATED PROTEIN 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PANCREATIC LIPASE RELATED PROTEIN 1 | | Authors: | Roussel, A, Cambillau, C. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reactivation of the totally inactive pancreatic lipase RP1 by structure-predicted point mutations.
Proteins, 32, 1998
|
|
1RL0
 
 | | Crystal structure of a new ribosome-inactivating protein (RIP): dianthin 30 | | Descriptor: | Antiviral protein DAP-30 | | Authors: | Fermani, S, Falini, G, Ripamonti, A, Bolognesi, A, Polito, L, Stirpe, F. | | Deposit date: | 2003-11-24 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.4A structure of dianthin 30 indicates a role of surface potential at the active site of type 1 ribosome inactivating proteins
J.Struct.Biol., 149, 2005
|
|
1RQN
 
 | | Phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
1RPX
 
 | |
1RTB
 
 | |
1RTF
 
 | |
1RS6
 
 | | Rat neuronal NOS heme domain with D-lysine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Neuronal and Endothelial Nitric Oxide Synthase Heme Domain with d-Nitroarginine-Containing Dipeptide Inhibitors Bound.
Biochemistry, 43, 2004
|
|
1RVB
 
 | |
1RLD
 
 | | SOLID-STATE PHASE TRANSITION IN THE CRYSTAL STRUCTURE OF RIBULOSE 1,5-BIPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE | | Descriptor: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Eisenberg, D. | | Deposit date: | 1993-12-10 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solid-state phase transition in the crystal structure of ribulose 1,5-bisphosphate carboxylase/oxygenase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
