1I90
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-8520 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 4-AMINO-3,4-DIHYDRO-2-(3-METHOXYPROPYL)-, 1,1-DIOXIDE, (R) | | Descriptor: | 4-AMINO-6-[N-(3-METHOXYLPROPYL)-2H-THIENO[3,2-E][1,2]THIAZINE 1,1-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
1RE4
 
 | | Crystal Structure of Fragment D of BbetaD398A Fibrinogen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha/alpha-E chain, ... | | Authors: | Kostelansky, M.S, Betts, L, Gorkun, O.V, Lord, S.T. | | Deposit date: | 2003-11-06 | | Release date: | 2004-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | BbetaGlu397 and BbetaAsp398 but not BbetaAsp432 are required for "B:b" interactions.
Biochemistry, 43, 2004
|
|
1I9O
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,3,4-TRIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(2,3,4-TRIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1I8Z
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-6629 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 2-(3-METHOXYPHENYL)-3-(4-MORPHOLINYL)-, 1,1-DIOXIDE | | Descriptor: | 6-[N-(3-METHOXY-PHENYL)-3-(MORPHOLIN-4-YLMETHYL)-2H-THIENO[3,2-E]-1,2-THIAZINE-1,1,-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
1I9Q
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(3,4,5-TRIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(3,4,5-TRIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1RH9
 
 | | Family GH5 endo-beta-mannanase from Lycopersicon esculentum (tomato) | | Descriptor: | endo-beta-mannanase | | Authors: | Oakley, A.J, Bourgault, R, Bewley, J.D, Wilce, M.C.J. | | Deposit date: | 2003-11-14 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure of (1,4)-beta-D-mannan mannanohydrolase from tomato fruit.
Protein Sci., 14, 2005
|
|
1RP9
 
 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) inactive mutant d180a in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase type 1 isozyme, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-12-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
1EYG
 
 | |
1RA0
 
 | | Bacterial cytosine deaminase D314G mutant bound to 5-fluoro-4-(S)-hydroxy-3,4-dihydropyrimidine. | | Descriptor: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1E5N
 
 | | E246C mutant of P fluorescens subsp. cellulosa xylanase A in complex with xylopentaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Lo Leggio, L, Jenkins, J.A, Harris, G.W, Pickersgill, R.W. | | Deposit date: | 2000-07-27 | | Release date: | 2000-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic study of xylopentaose binding to Pseudomonas fluorescens xylanase A.
Proteins, 41, 2000
|
|
1E60
 
 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES - Structure II BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-06 | | Release date: | 2000-08-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
1E7R
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase Y136E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1QH6
 
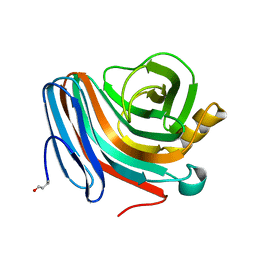 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
1QI3
 
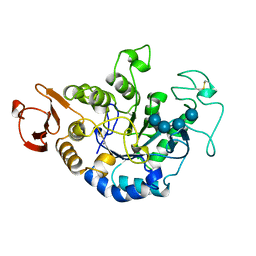 | | MUTANT (D193N) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (EXO-MALTOTETRAOHYDROLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hasegawa, K, Kubota, M, Matsuura, Y. | | Deposit date: | 1999-06-01 | | Release date: | 1999-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
1QIR
 
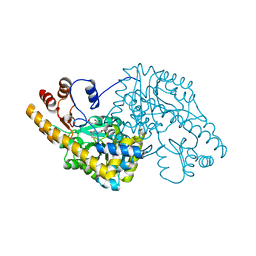 | | ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI, C191Y MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of Residues Outside the Active Site in Catalysis: Structural Basis for Function of C191 Mutants of E. Coli Aspartate Aminotransferase
Protein Eng., 13, 2000
|
|
1QJC
 
 | |
1EF7
 
 | | CRYSTAL STRUCTURE OF HUMAN CATHEPSIN X | | Descriptor: | CATHEPSIN X | | Authors: | Guncar, G, Klemencic, I, Turk, B, Turk, V, Karaoglanovic-Carmona, A, Juliano, L, Turk, D. | | Deposit date: | 2000-02-07 | | Release date: | 2000-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of cathepsin X: a flip-flop of the ring of His23 allows carboxy-monopeptidase and carboxy-dipeptidase activity of the protease.
Structure Fold.Des., 8, 2000
|
|
1E61
 
 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES - Structure II BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-06 | | Release date: | 2000-08-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
1E6X
 
 | | MYROSINASE FROM SINAPIS ALBA with a bound transition state analogue,D-glucono-1,5-lactone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-glucono-1,5-lactone, ... | | Authors: | Burmeister, W.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-01-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High Resolution X-Ray Crystallography Shows that Ascorbate is a Cofactor for Myrosinase and Substitutes for the Function of the Catalytic Base
J.Biol.Chem., 275, 2000
|
|
1QIP
 
 | | HUMAN GLYOXALASE I COMPLEXED WITH S-P-NITROBENZYLOXYCARBONYLGLUTATHIONE | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (LACTOYLGLUTATHIONE LYASE), S-P-NITROBENZYLOXYCARBONYLGLUTATHIONE, ... | | Authors: | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | Deposit date: | 1999-06-14 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Reaction mechanism of glyoxalase I explored by an X-ray crystallographic analysis of the human enzyme in complex with a transition state analogue.
Biochemistry, 38, 1999
|
|
1ENX
 
 | |
1EEZ
 
 | |
1Q91
 
 | | Crystal structure of human mitochondrial deoxyribonucleotidase in complex with the inhibitor DPB-T | | Descriptor: | (S)-1-[2'-DEOXY-3',5'-O-(1-PHOSPHONO)BENZYLIDENE-B-D-THREO-PENTOFURANOSYL]THYMINE, 5(3)-deoxyribonucleotidase, MAGNESIUM ION, ... | | Authors: | Rinaldo-Matthis, A, Rampazzo, C, Balzarini, J, Reichard, P, Bianchi, V, Nordlund, P. | | Deposit date: | 2003-08-22 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the mitochondrial deoxyribonucleotidase in complex with two specific inhibitors
Mol.Pharmacol., 65, 2004
|
|
1Q9F
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX.
J.Mol.Biol., 336, 2004
|
|
1Q8W
 
 | | The Catalytic Subunit of cAMP-dependent Protein Kinase in Complex with Rho-kinase Inhibitor Fasudil (HA-1077) | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C, Gassel, M, Hidaka, H, Kinzel, V, Huber, R, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2003-08-22 | | Release date: | 2003-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein kinase A in complex with Rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: structural basis of selectivity.
Structure, 11, 2003
|
|
