1GQF
 
 | |
2CWA
 
 | |
8FHY
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with neutralizing antibody WRAIR-5021 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MALONATE ION, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2022-12-15 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Diverse array of neutralizing antibodies elicited upon Spike Ferritin Nanoparticle vaccination in rhesus macaques.
Nat Commun, 15, 2024
|
|
1GVG
 
 | | Crystal Structure of Clavaminate Synthase with Nitric Oxide | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, DEOXYGUANIDINOPROCLAVAMINIC ACID, ... | | Authors: | Zhang, Z.H, Ren, J, McKinnon, C.H, Clifton, I.J, Harlos, K, Schofield, C.J. | | Deposit date: | 2002-02-12 | | Release date: | 2003-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal Structure of a Clavaminate Synthase-Fe(II) -2-Oxoglutarate-Substrate-No Complex: Evidence for Metal Centered Rearrangements
FEBS Lett., 517, 2002
|
|
2CX3
 
 | | Crystal structure of a bacterioferritin comigratory protein peroxiredoxin from the Aeropyrum pernix K1 (form-1 crystal) | | Descriptor: | bacterioferritin comigratory protein | | Authors: | Mizohata, E, Murayama, K, Idaka, M, Tatsuguchi, A, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-27 | | Release date: | 2005-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of a bacterioferritin comigratory protein peroxiredoxin from the Aeropyrum pernix K1 (form-1 crystal)
To be Published
|
|
2CXH
 
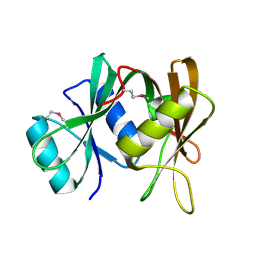 | | Crystal structure of probable ribosomal biogenesis protein from Aeropyrum pernix K1 | | Descriptor: | Probable brix-domain ribosomal biogenesis protein | | Authors: | Kawazoe, M, Takemoto, C, Hanawa-Suetsugu, K, Kaminishi, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-29 | | Release date: | 2005-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of probable ribosomal biogenesis protein from Aeropyrum pernix K1
To be Published
|
|
1GYG
 
 | |
1H03
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
2CKX
 
 | |
1GT4
 
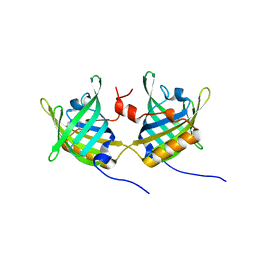 | | Complex of Bovine Odorant Binding Protein with undecanal | | Descriptor: | ODORANT-BINDING PROTEIN, UNDECANAL | | Authors: | Vincent, F, Ramoni, R, Spinelli, S, Grolli, S, Conti, V, Cambillau, C, Tegoni, M. | | Deposit date: | 2002-01-10 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Bovine Odorant-Binding Protein in Complex with Odorant Molecules.
Eur.J.Biochem., 271, 2004
|
|
1H1T
 
 | |
4Y1N
 
 | | Oceanobacillus iheyensis group II intron domain 1 with iridium hexamine | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Zhao, C, Rajashankar, K.R, Marcia, M, Pyle, A.M. | | Deposit date: | 2015-02-08 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of group II intron domain 1 reveals a template for RNA assembly.
Nat.Chem.Biol., 11, 2015
|
|
1GPF
 
 | |
2CZD
 
 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution
To be Published
|
|
1H23
 
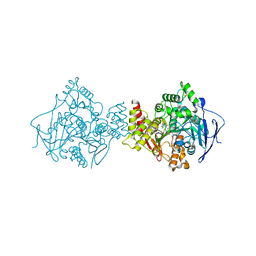 | | Structure of acetylcholinesterase (E.C. 3.1.1.7) complexed with (S,S)-(-)-bis(12)-hupyridone at 2.15A resolution | | Descriptor: | (S,S)-(-)-N,N'-DI-5'-[5',6',7',8'-TETRAHYDRO- 2'(1'H)-QUINOLYNYL]-1,12-DIAMINODODECANE DIHYDROCHLORIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Wong, D.M, Greenblatt, H.M, Carlier, P.R, Han, Y.F, Pang, Y.P, Silman, I, Sussman, J.L. | | Deposit date: | 2002-07-30 | | Release date: | 2002-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Acetylcholinesterase Complexed with Bivalent Ligands Related to Huperzine A: Experimental Evidence for Species-Dependent Protein-Ligand Complementarity
J.Am.Chem.Soc., 125, 2003
|
|
2CO3
 
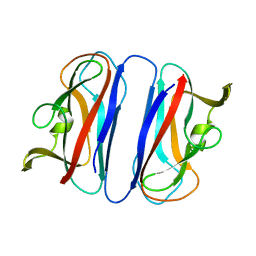 | | Salmonella enterica SafA pilin, head-to-tail swapped dimer of Ntd1 mutant | | Descriptor: | SAFA PILUS SUBUNIT | | Authors: | Remaut, H, Rose, R.J, Hannan, T.J, Hultgren, S.J, Radford, S.E, Ashcroft, A.E, Waksman, G. | | Deposit date: | 2006-05-25 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Donor-Strand Exchange in Chaperone-Assisted Pilus Assembly Proceeds Through a Concerted Beta-Strand Displacement Mechanism
Mol.Cell, 22, 2006
|
|
1GRQ
 
 | |
1H2F
 
 | |
1GSL
 
 | |
1H43
 
 | | R210E N-TERMINAL LOBE HUMAN LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Peterson, N.A, Arcus, V.L, Anderson, B.F, Jameson, G.B, Tweedie, J.W, Baker, E.N. | | Deposit date: | 2002-10-02 | | Release date: | 2002-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | "Dilysine Trigger" in Transferrins Probed by Mutagenesis of Lactoferrin: Crystal Structures of the R210G, R210E, and R210L Mutants of Human Lactoferrin
Biochemistry, 41, 2002
|
|
1GVN
 
 | | Crystal Structure of the Plasmid Maintenance System epsilon/zeta: Meachnism of toxin inactivation and toxin function | | Descriptor: | EPSILON, SULFATE ION, ZETA | | Authors: | Meinhart, A, Alonso, J.C, Straeter, N, Saenger, W. | | Deposit date: | 2002-02-19 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Plasmid Maintenance System Epsilon /Zeta : Functional Mechanism of Toxin Zeta and Inactivation by Epsilon 2 Zeta 2 Complex Formation
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1H55
 
 | | STRUCTURE OF HORSERADISH PEROXIDASE C1A COMPOUND II | | Descriptor: | ACETATE ION, CALCIUM ION, OXYGEN ATOM, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-18 | | Release date: | 2002-06-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
4Y75
 
 | | Yeast 20S proteasome in complex with Ac-PAF-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PAF-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
1H5K
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (78-89% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H04
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, NICKEL (II) ION | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
