3LN2
 
 | |
3LN9
 
 | | Crystal structure of the fibril-specific B10 antibody fragment | | Descriptor: | CITRATE ANION, GLYCEROL, Immunoglobulin heavy chain antibody variable domain B10, ... | | Authors: | Parthier, C, Morgado, I, Stubbs, M.T, Faendrich, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Amyloid Fibril Recognition with the Conformational B10 Antibody Fragment Depends on Electrostatic Interactions.
J.Mol.Biol., 2010
|
|
5HVC
 
 | | Solution structure of the apo state of the acyl carrier protein from the MLSA2 subunit of the mycolactone polyketide synthase | | Descriptor: | Type I modular polyketide synthase | | Authors: | Vance, S, Tkachenko, O, Thomas, B, Bassuni, M, Hong, H, Nietlispach, D, Broadhurst, R.W. | | Deposit date: | 2016-01-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sticky swinging arm dynamics: studies of an acyl carrier protein domain from the mycolactone polyketide synthase.
Biochem.J., 473, 2016
|
|
5H5V
 
 | |
3LB6
 
 | | The structure of IL-13 in complex with IL-13Ralpha2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Interleukin-13, ... | | Authors: | Lupardus, P.J, Garcia, K.C, Birnbaum, M.E. | | Deposit date: | 2010-01-07 | | Release date: | 2010-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular basis for shared cytokine recognition revealed in the structure of an unusually high affinity complex between IL-13 and IL-13Ralpha2.
Structure, 18, 2010
|
|
3LNE
 
 | |
5H7B
 
 | | Crystal structure of a repeat protein with five Protein A repeat modules | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H7N
 
 | | Crystal structure of human NLRP12-PYD with a MBP tag | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NLRP12-PYD with MBP tag, ... | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-19 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
3LC8
 
 | |
3LNN
 
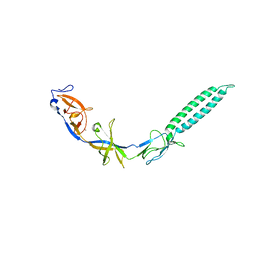 | | Crystal structure of ZneB from Cupriavidus metallidurans | | Descriptor: | Membrane fusion protein (MFP) heavy metal cation efflux ZneB (CzcB-like), ZINC ION | | Authors: | Lee, J.K, De Angelis, F, Miercke, L.J, Stroud, R.M, Vandenbussche, G, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-02-02 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5H7Z
 
 | |
5H89
 
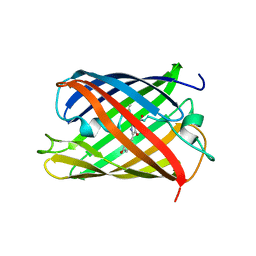 | | Crystal structure of mRojoA mutant - T16V - P63Y - W143G - L163V | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
3LCT
 
 | |
5H8L
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) C158S mutant in complex with putrescine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
3L61
 
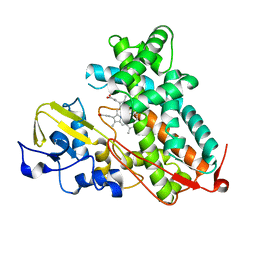 | | Crystal structure of substrate-free P450cam at 200 mM [K+] | | Descriptor: | Camphor 5-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, Y.-T, Wilson, R.F, Rupniewski, I, Goodin, D.B. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | P450cam visits an open conformation in the absence of substrate.
Biochemistry, 49, 2010
|
|
5HAA
 
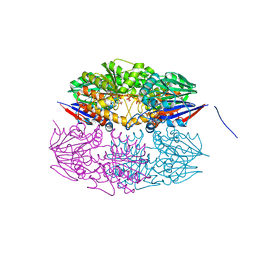 | |
3LD7
 
 | | Crystal structure of the Lin0431 protein from Listeria innocua, Northeast Structural Genomics Consortium Target LkR112 | | Descriptor: | Lin0431 protein | | Authors: | Vorobiev, S, Chen, Y, Lee, D, Patel, D.J, Ciccosanti, C, Sahdev, S, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-12 | | Release date: | 2010-01-26 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Crystal structure of the Lin0431 protein from Listeria innocua
To be Published
|
|
5HAY
 
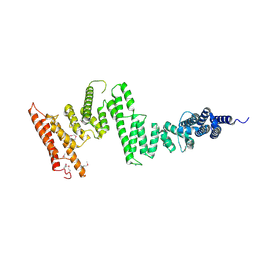 | |
6OLT
 
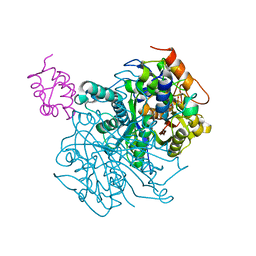 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C12-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, N-[2-(dodecanoylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Mindrebo, J.T, Kim, W.E, Bartholow, T.G, Chen, A, Davis, T.D, La Clair, J, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-04-17 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Gating mechanism of elongating beta-ketoacyl-ACP synthases.
Nat Commun, 11, 2020
|
|
5HBC
 
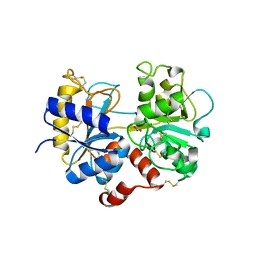 | | Intermediate structure of iron-saturated C-lobe of bovine lactoferrin at 2.79 Angstrom resolution indicates the softening of iron coordination | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, FE (III) ION, ... | | Authors: | Singh, A, Rastogi, N, Singh, P.K, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-12-31 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of iron saturated C-lobe of bovine lactoferrin at pH 6.8 indicates a weakening of iron coordination
Proteins, 84, 2016
|
|
3L6F
 
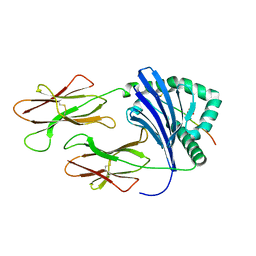 | |
3LDF
 
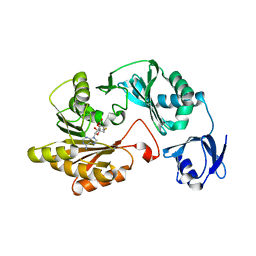 | |
5HBL
 
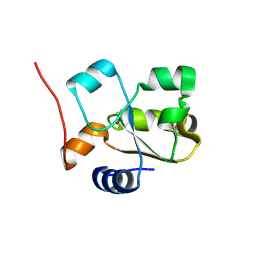 | | Native rhodanese domain of YgaP prepared with 1mM DDT is S-nitrosylated | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2015-12-31 | | Release date: | 2016-08-10 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5H60
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | MANGANESE (II) ION, Transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-20 | | Last modified: | 2018-10-31 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
3LDU
 
 | | The crystal structure of a possible methylase from Clostridium difficile 630. | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tan, K, Wu, R, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a possible methylase from Clostridium difficile 630.
To be Published
|
|
