8J8L
 
 | | Overall structure of the LAT1-4F2hc bound with L-dopa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4-DIHYDROXYPHENYLALANINE, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Yan, R.H, Li, Y.N, Shi, T.H. | | Deposit date: | 2023-05-02 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Overall structure of the LAT1-4F2hc bound with L-dopa
To Be Published
|
|
8J8M
 
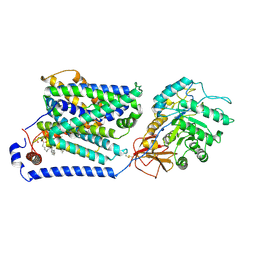 | | Overall structure of the LAT1-4F2hc bound with tryptophan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Large neutral amino acids transporter small subunit 1, ... | | Authors: | Yan, R.H, Li, Y.N, Shi, T.H. | | Deposit date: | 2023-05-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Overall structure of the LAT1-4F2hc bound with tryptophan
To Be Published
|
|
8JGK
 
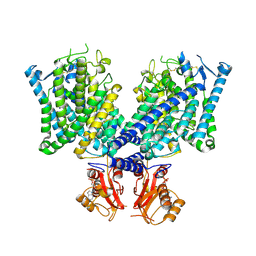 | | Cryo-EM structure of mClC-3 with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
8JEV
 
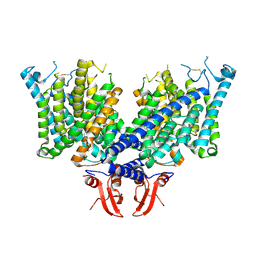 | | Cryo-EM structure of apo state mClC-3 | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-16 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
8JGV
 
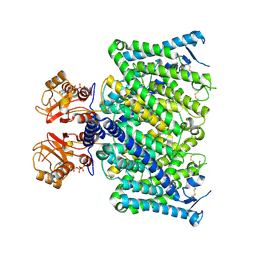 | | Cryo-EM structure of mClC-3_I607T with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
8JGJ
 
 | | Cryo-EM structure of mClC-3 with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
1GWW
 
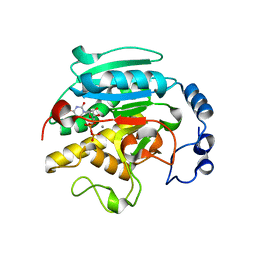 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - ALPHA-D-GLUCOSE COMPLEX | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
8JGS
 
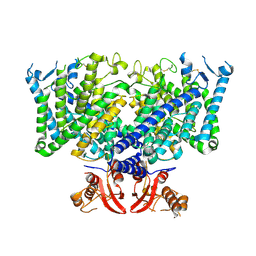 | | Cryo-EM structure of apo state mClC-3_I607T | | Descriptor: | H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
3O6M
 
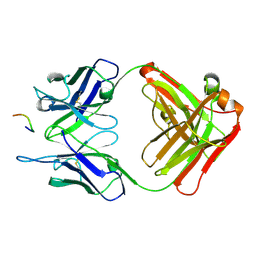 | | Anti-Tat HIV 11H6H1 Fab' complexed with a 9-mer Tat peptide | | Descriptor: | 11H6H1 Fab' heavy chain, 11H6H1 Fab' light chain, Protein Tat 9-mer peptide | | Authors: | Serriere, J, Gouet, P, Guillon, C. | | Deposit date: | 2010-07-29 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fab'-induced folding of antigenic N-terminal peptides from intrinsically disordered HIV-1 Tat revealed by X-ray crystallography.
J.Mol.Biol., 405, 2011
|
|
8JGL
 
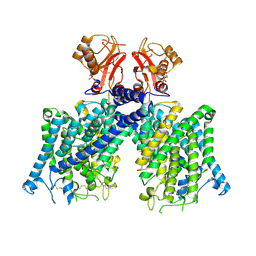 | | Cryo-EM structure of mClC-3 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-21 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
1HDK
 
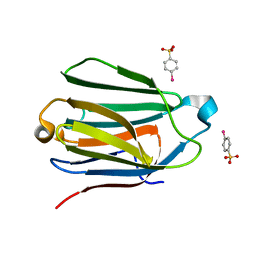 | | Charcot-Leyden Crystal Protein - pCMBS Complex | | Descriptor: | EOSINOPHIL LYSOPHOSPHOLIPASE, PARA-MERCURY-BENZENESULFONIC ACID | | Authors: | Ackerman, S.J, Savage, M.P, Liu, L, Leonidas, D.D, Kwatia, M.A, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Charcot-Leyden Crystal Protein (Galectin-10) is not a Dual Function Galectin with Lysophospholipase Activity But Binds a Lysophospholipase Inhibitor in a Novel Structural Fashion.
J.Biol.Chem., 277, 2002
|
|
1HF6
 
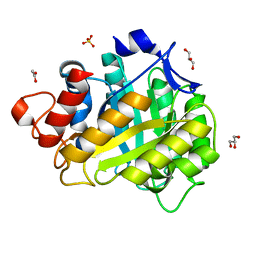 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE | | Descriptor: | ACETIC ACID, ENDOGLUCANASE B, GLYCEROL, ... | | Authors: | Varrot, A, Withers, S, Vasella, A, Schulein, M, Davies, G.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Direct Experimental Observation of the Hydrogen-Bonding Network of a Glycosidase Along its Reaction Coordinate Revealed by Atomic Resolution Analyses of Endoglucanase Cel5A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1H36
 
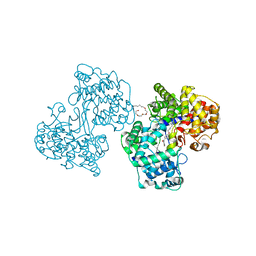 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (4-BROMOPHENYL)[4-({(2E)-4-[CYCLOPROPYL(METHYL)AMINO]BUT-2-ENYL}OXY)PHENYL]METHANONE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
8KA5
 
 | |
8JL3
 
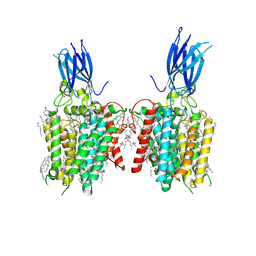 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure and mechanism of lysosome transmembrane acetylation by HGSNAT.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8KEX
 
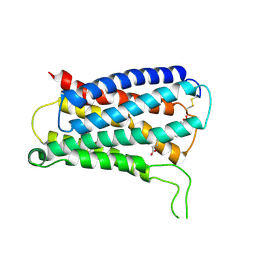 | | CryoEM structure of Gq coupled MRGPRX4 with agonist DCA-3P, local | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},12~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-12-oxidanyl-3-phosphonooxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Soluble cytochrome b562,Mas-related G-protein coupled receptor member X4,Green fluorescent protein | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2023-08-14 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-guided discovery of bile acid derivatives for treating liver diseases without causing itch.
Cell, 2024
|
|
8K2W
 
 | | Structure of CXCR3 complexed with antagonist AMG487 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, N-[(1R)-1-[3-(4-ethoxyphenyl)-4-oxidanylidene-pyrido[2,3-d]pyrimidin-2-yl]ethyl]-N-(pyridin-3-ylmethyl)-2-[4-(trifluoromethyloxy)phenyl]ethanamide, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure basis for the modulation of CXC chemokine receptor 3 by antagonist AMG487.
Cell Discov, 9, 2023
|
|
8KA3
 
 | |
1GQK
 
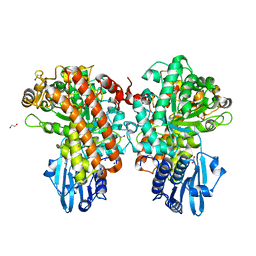 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1GR7
 
 | | Crystal structure of the double mutant Cys3Ser/Ser100Pro from Pseudomonas Aeruginosa at 1.8 A resolution | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Okvist, M, Bonander, N, Sandberg, A, Karlsson, B.G, Krengel, U, Xue, Y, Sjolin, L. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the double azurin mutant Cys3Ser/Ser100Pro from Pseudomonas aeruginosa at 1.8 A resolution: its folding-unfolding energy and unfolding kinetics.
Biochim.Biophys.Acta, 1596, 2002
|
|
8JPR
 
 | | Cryo-EM structure of Y553C human ClC-6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, H(+)/Cl(-) exchange transporter 6, ... | | Authors: | Zhang, S.S. | | Deposit date: | 2023-06-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of ClC-6 function and its impairment in human disease
Sci Adv, 2023
|
|
1OGB
 
 | | Chitinase b from Serratia marcescens mutant D142N | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Vaaje-Kolstad, G, Houston, D.R, Rao, F.V, Peter, M.G, Synstad, B, van Aalten, D.M.F, Eijsink, V.G.H. | | Deposit date: | 2003-04-29 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the D142N Mutant of the Family 18 Chitinase Chib from Serratia Marcescens and its Complex with Allosamidin
Biochim.Biophys.Acta, 1696, 2004
|
|
8JMT
 
 | | Structure of the adhesion GPCR ADGRL3 in the apo state | | Descriptor: | Adhesion G protein-coupled receptor L3,Soluble cytochrome b562 | | Authors: | Tao, Y, Guo, Q, He, B, Zhong, Y. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8JL4
 
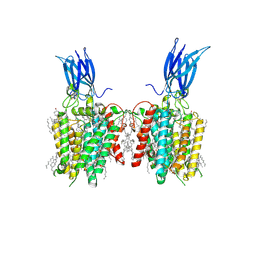 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | membrane proteins
To Be Published
|
|
8JLX
 
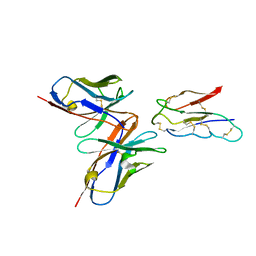 | | CCHFV envelope protein Gc in complex with Gc13 | | Descriptor: | Glycoprotein C,CCHFV Gc fusion loops, Mouse antibody Gc13 heavy chain, Mouse antibody Gc13 light chain | | Authors: | Chong, T, Cao, S. | | Deposit date: | 2023-06-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralizing monoclonal antibodies against the Gc fusion loop region of Crimean-Congo hemorrhagic fever virus.
Plos Pathog., 20, 2024
|
|
