6Q4B
 
 | | CDK2 in complex with FragLite13 | | Descriptor: | 5-bromanylpyrimidine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4J
 
 | | CDK2 in complex with FragLite34 | | Descriptor: | 2-[3-(pyrimidin-4-ylamino)phenyl]ethanoic acid, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
5Y6I
 
 | |
5UQ3
 
 | |
6Q3C
 
 | | CDK2 in complex with FragLite1 | | Descriptor: | 4-bromo-1H-pyrazole, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q3B
 
 | | CDK2 in complex with FragLite2 | | Descriptor: | 4-IODOPYRAZOLE, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q49
 
 | | CDK2 in complex with FragLite6 | | Descriptor: | 4-bromanyl-1~{H}-pyridin-2-one, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4G
 
 | | CDK2 in complex with FragLite37 | | Descriptor: | 2-[3-(2-azanyl-9~{H}-purin-6-yl)phenyl]ethanoic acid, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
7EGM
 
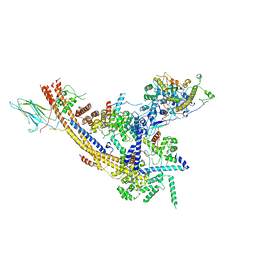 | | The SRM module of SWI/SNF-nucleosome complex | | Descriptor: | SWI/SNF chromatin-remodeling complex subunit SNF5, SWI/SNF chromatin-remodeling complex subunit SWI1, SWI/SNF complex subunit SWI3, ... | | Authors: | Chen, Z.C, Chen, K.J, He, Z.Y, Ye, Y.P. | | Deposit date: | 2021-03-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the SWI/SNF complex bound to the nucleosome and insights into the functional modularity.
Cell Discov, 7, 2021
|
|
6Q48
 
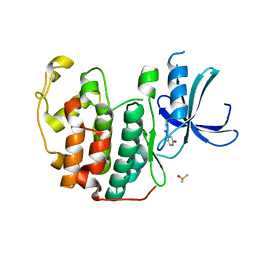 | | CDK2 in complex with FragLite7 | | Descriptor: | 4-iodanyl-3~{H}-pyridin-2-one, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4H
 
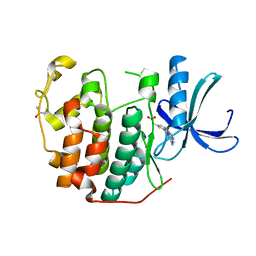 | | CDK2 in complex with FragLite36 | | Descriptor: | 2-[3-[(2-azanyl-9~{H}-purin-6-yl)oxy]phenyl]ethanoic acid, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
8DJH
 
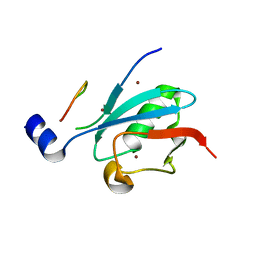 | | Ternary complex of SUMO1 with a phosphomimetic SIM of PML and zinc | | Descriptor: | PML 4SD, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Bourdeau, V, Gagnon, C, Igelmann, S, Sakaguchi, K, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1.
Nucleic Acids Res., 50, 2022
|
|
6BY1
 
 | | E. coli pH03H9 complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Amiri, H, Noller, H.F. | | Deposit date: | 2017-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Structural evidence for product stabilization by the ribosomal mRNA helicase.
Rna, 25, 2019
|
|
8DJI
 
 | | Ternary complex of SUMO1 with the SIM of PML and zinc | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Bourdeau, V, Gagnon, C, Igelmann, S, Sakaguchi, K, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1.
Nucleic Acids Res., 50, 2022
|
|
1A1D
 
 | | YEAST RNA POLYMERASE SUBUNIT RPB8, NMR, MINIMIZED AVERAGE STRUCTURE, ALPHA CARBONS ONLY | | Descriptor: | RNA POLYMERASE | | Authors: | Krapp, S, Kelly, G, Reischl, J, Weinzierl, R, Matthews, S. | | Deposit date: | 1997-12-10 | | Release date: | 1999-03-02 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic RNA polymerase subunit RPB8 is a new relative of the OB family.
Nat.Struct.Biol., 5, 1998
|
|
6CQH
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N-propynyl-N-ethyl-dihydropteridine inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-8-ethyl-7-methyl-5-(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-15 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N-propynyl-N-ethyl-dihydropteridine inhibitor
To Be Published
|
|
5LQW
 
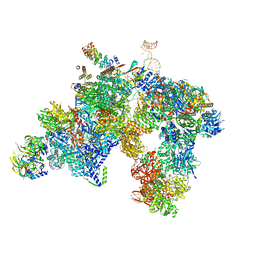 | | yeast activated spliceosome | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-processing protein 45, Pre-mRNA-splicing factor 8, ... | | Authors: | Rauhut, R, Luehrmann, R. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Molecular architecture of the Saccharomyces cerevisiae activated spliceosome
Science, 6306, 2016
|
|
6CMM
 
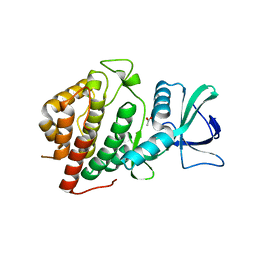 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-05 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor
To Be Published
|
|
6D36
 
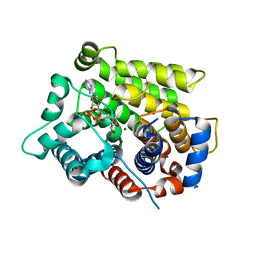 | | Structure of human ARH3 bound to ADP-ribose and magnesium | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pourfarjam, Y, Ventura, J, Kurinov, I, Kim, I.K. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition.
J.Biol.Chem., 293, 2018
|
|
6D3A
 
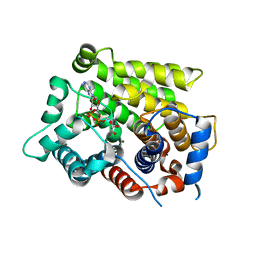 | | Structure of human ARH3 D314E bound to ADP-ribose and magnesium | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pourfarjam, Y, Ventura, J, Kurinov, I, Kim, I.K. | | Deposit date: | 2018-04-15 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.60001016 Å) | | Cite: | Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition.
J.Biol.Chem., 293, 2018
|
|
6CSW
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N-methyl-N-propyl-dihydropteridine inhibitor | | Descriptor: | (7R)-5-butyl-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7,8-dimethyl-7,8-dihydropteridin-6(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Chiodi, C.G, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-21 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N-methyl-N-propyl-dihydropteridine inhibitor
To Be Published
|
|
3EGC
 
 | | Crystal structure of a putative ribose operon repressor from Burkholderia thailandensis | | Descriptor: | putative ribose operon repressor | | Authors: | Bonanno, J.B, Patskovsky, Y, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a putative ribose operon repressor from Burkholderia thailandensis
To be Published
|
|
3ER6
 
 | | Crystal structure of a putative transcriptional regulator protein from Vibrio parahaemolyticus | | Descriptor: | Putative transcriptional regulator protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative transcriptional regulator protein from Vibrio parahaemolyticus
To be Published
|
|
3E2B
 
 | | Crystal structure of Dynein Light chain LC8 in complex with a peptide derived from Swallow | | Descriptor: | ACETATE ION, Dynein light chain 1, cytoplasmic, ... | | Authors: | Benison, G, Barbar, E, Karplus, P.A. | | Deposit date: | 2008-08-05 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The interplay of ligand binding and quaternary structure in the diverse interactions of dynein light chain LC8.
J.Mol.Biol., 384, 2008
|
|
3E2R
 
 | |
