9DVJ
 
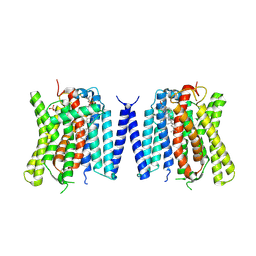 | | "Structure of the phosphate exporter XPR1/SLC53A1 | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, PHOSPHATE ION, ... | | Authors: | Zhu, Q, Diver, M.M. | | Deposit date: | 2024-10-08 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Transport and InsP 8 gating mechanisms of the human inorganic phosphate exporter XPR1.
Nat Commun, 16, 2025
|
|
4W9G
 
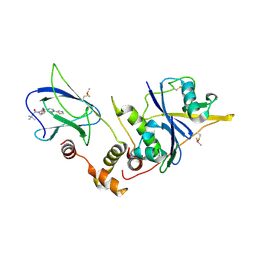 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(3-methyl-4-(thiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 6) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[3-methyl-4-(1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
8VV8
 
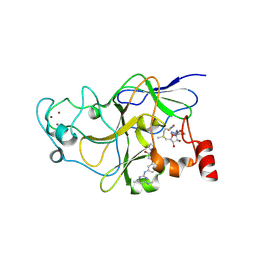 | | Crystal Structure of EHMT2 bound to EZM8266 | | Descriptor: | (2R)-1-(azetidin-1-yl)-3-(2-methoxy-5-{[4-methyl-6-(methylamino)pyrimidin-2-yl]amino}phenoxy)propan-2-ol, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Cocozaki, A, Farrow, N.A. | | Deposit date: | 2024-01-30 | | Release date: | 2024-12-11 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combining EHMT and PARP Inhibition: A Strategy to Diminish Therapy-Resistant Ovarian Cancer Tumor Growth while Stimulating Immune Activation.
Mol.Cancer Ther., 23, 2024
|
|
8W38
 
 | | TAS-120 covalent structure with FGFR2 molecular brake mutant | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 2, SULFATE ION | | Authors: | Hoffman, I.D, Nelson, K.J, Bensen, D.C, Bailey, J.B. | | Deposit date: | 2024-02-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A model for decoding resistance in precision oncology: acquired resistance to FGFR inhibitors in cholangiocarcinoma.
Ann Oncol, 36, 2025
|
|
8Y0L
 
 | |
1G27
 
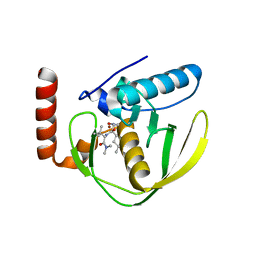 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | Descriptor: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-17 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
8Y0M
 
 | |
6C6B
 
 | |
9BUF
 
 | |
9FTT
 
 | |
9FX0
 
 | |
9FW9
 
 | | Cryo-EM structure of the type 1 pilus assembly platform as part of the FimA-bound chaperone-usher pilus complex (FimDHGFAnC - body 2) | | Descriptor: | Chaperone protein FimC, Outer membrane usher protein FimD, Type-1 fimbrial protein, ... | | Authors: | Bachmann, P, Afanasyev, P, Boehringer, D, Glockshuber, R. | | Deposit date: | 2024-06-28 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the type 1 pilus assembly platform as part of the FimA-bound chaperone-usher pilus complex (FimDHGFAnC - body 2)
To Be Published
|
|
6HCN
 
 | | Adenovirus Type 5 Fiber Knob protein at 1.49A resolution | | Descriptor: | 1,2-ETHANEDIOL, Fiber protein, MAGNESIUM ION | | Authors: | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | Deposit date: | 2018-08-15 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions.
Nat Commun, 10, 2019
|
|
9CPI
 
 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-[1-(2-aminoethyl)-4,5-dihydro-1H-imidazol-2-yl]-N-[4-(4,5-dihydro-1H-imidazol-2-yl)phenyl]benzamide, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | NMR structures of small molecules bound to a model of a CUG RNA repeat expansion.
Bioorg.Med.Chem.Lett., 111, 2024
|
|
8BR6
 
 | | Discovery of IRAK4 Inhibitor 40 | | Descriptor: | ACETATE ION, Interleukin-1 receptor-associated kinase 4, ~{N}-[6-methoxy-2-(2-methylsulfonylethyl)-1,3-dihydroindazol-5-yl]-6-(2-oxidanylpropan-2-yl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wengner, A.M, Guimond, N, Thaler, T, Platzek, J, Ewerspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-11-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.167 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
8SDV
 
 | | Crystal structure of PDC-3 Y221H beta-lactamase in complex with the boronic acid inhibitor S02030 | | Descriptor: | 1-{(2R)-2-(dihydroxyboranyl)-2-[(thiophen-2-ylacetyl)amino]ethyl}-1H-1,2,3-triazole-4-carboxylic acid, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Natural protein engineering in the Omega-loop: the role of Y221 in ceftazidime and ceftolozane resistance in Pseudomonas -derived cephalosporinase.
Antimicrob.Agents Chemother., 67, 2023
|
|
9FO4
 
 | | Half-closed CODH/ACS (Class 2) in the methylated state | | Descriptor: | CO-dehydrogenase, CO-methylating acetyl-CoA synthase, Fe(3)-Ni(1)-S(4) cluster, ... | | Authors: | Ruickoldt, J, Wendler, P, Dobbek, H. | | Deposit date: | 2024-06-11 | | Release date: | 2025-06-25 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Ligand binding to a Ni-Fe cluster orchestrates conformational changes of the CO-dehydrogenase-acetyl-CoA synthase complex.
Nat Catal, 8, 2025
|
|
6T71
 
 | |
8BR7
 
 | | Discovery of IRAK4 Inhibitors BAY1834845 and BAY1830839 | | Descriptor: | 3-nitro-~{N}-[2-[2-oxidanylidene-2-[4-(phenylcarbonyl)piperazin-1-yl]ethyl]indazol-5-yl]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-11-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
8SDT
 
 | |
9FOX
 
 | | Half-closed CODH/ACS in the reduced state | | Descriptor: | CO-dehydrogenase, CO-methylating acetyl-CoA synthase, Fe(3)-Ni(1)-S(4) cluster, ... | | Authors: | Ruickoldt, J, Wendler, P, Dobbek, H. | | Deposit date: | 2024-06-12 | | Release date: | 2025-06-25 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Ligand binding to a Ni-Fe cluster orchestrates conformational changes of the CO-dehydrogenase-acetyl-CoA synthase complex.
Nat Catal, 8, 2025
|
|
8YHI
 
 | | The Crystal Structure of SHP1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Shen, Z. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of SHP1 from Biortus.
To Be Published
|
|
7AF1
 
 | | The structure of Artemis/SNM1C/DCLRE1C with 2 Zinc ions | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
5KPR
 
 | | PANK3-AMPPNP-Pantothenate complex | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PANTOTHENOIC ACID, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2016-07-05 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | Allosteric Regulation of Mammalian Pantothenate Kinase.
J.Biol.Chem., 291, 2016
|
|
8SC3
 
 | |
