1TE2
 
 | | Putative Phosphatase Ynic from Escherichia coli K12 | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 2-deoxyglucose-6-P phosphatase, CALCIUM ION | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Putative Phosphatase Ynic from Escherichia coli K12
To be Published
|
|
1TE3
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | CHLORIDE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-24 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1TE4
 
 | | Solution structure of MTH187. Ontario Centre for Structural Proteomics target MTH0187_1_111; Northeast Structural Genomics Target TT740 | | Descriptor: | conserved protein MTH187 | | Authors: | Gignac, I, Julien, O, Yee, A, Arrowsmith, C.H, Gagne, S.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-05-24 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | MTH187 from Methanobacterium thermoautotrophicum has three HEAT-like Repeats.
J.Biomol.Nmr, 35, 2006
|
|
1TE5
 
 | |
1TE6
 
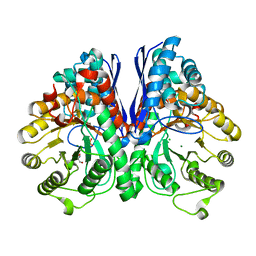 | | Crystal Structure of Human Neuron Specific Enolase at 1.8 angstrom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Gamma enolase, ... | | Authors: | Chai, G, Brewer, J, Lovelace, L, Aoki, T, Minor, W, Lebioda, L. | | Deposit date: | 2004-05-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expression, Purification and the 1.8 A Resolution Crystal Structure of Human Neuron Specific Enolase
J.Mol.Biol., 341, 2004
|
|
1TE7
 
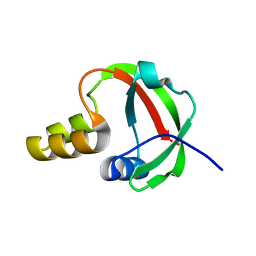 | | Solution NMR Structure of Protein yqfB from Escherichia coli. Northeast Structural Genomics Consortium Target ET99 | | Descriptor: | Hypothetical UPF0267 protein yqfB | | Authors: | Atreya, H.S, Shen, Y, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-05-24 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | G-Matrix Fourier Transform NOESY-Based Protocol for High-Quality Protein Structure Determination
J.Am.Chem.Soc., 127, 2005
|
|
1TEC
 
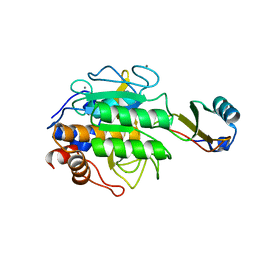 | | CRYSTALLOGRAPHIC REFINEMENT BY INCORPORATION OF MOLECULAR DYNAMICS. THE THERMOSTABLE SERINE PROTEASE THERMITASE COMPLEXED WITH EGLIN-C | | Descriptor: | CALCIUM ION, EGLIN C, SODIUM ION, ... | | Authors: | Gros, P, Dijkstra, B.W, Hol, W.G.J. | | Deposit date: | 1989-05-24 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic refinement by incorporation of molecular dynamics: thermostable serine protease thermitase complexed with eglin c.
Acta Crystallogr.,Sect.B, 45, 1989
|
|
1TED
 
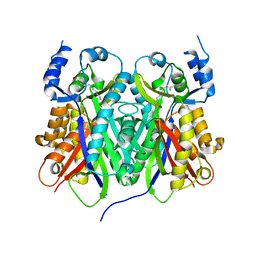 | |
1TEE
 
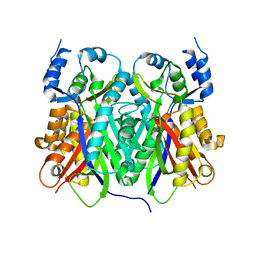 | |
1TEF
 
 | | Crystal structure of the spinach plastocyanin mutants G8D/K30C/T69C and K30C/T69C- a study of the effect on crystal packing and thermostability from the introduction of a novel disulfide bond | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Plastocyanin, ... | | Authors: | Okvist, M, Jacobson, F, Jansson, H, Hansson, O, Sjolin, L. | | Deposit date: | 2004-05-25 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel Disulfide Bonds Effect the Thermostability of Plastocyanin. Crystal structures of the triple plastocyanin mutant G8D/K30C/T69C and the double plastocyanin mutant K30C/T69C from spinach at 1.90 and 1.96 resolution, respectively.
To be Published
|
|
1TEG
 
 | | Crystal structure of the spinach plastocyanin mutants G8D/K30C/T69C and K30C/T69C- a study of the effect on crystal packing and thermostability from the introduction of a novel disulfide bond | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Plastocyanin, ... | | Authors: | Okvist, M, Jacobson, F, Jansson, H, Hansson, O, Sjolin, L. | | Deposit date: | 2004-05-25 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Novel Disulfide Bonds Effect the Thermostability of Plastocyanin. Crystal structures of the triple plastocyanin
mutant G8D/K30C/T69C and the double plastocyanin mutant K30C/T69C from spinach at 1.90 A and 1.96 A resolution, respectively.
To be Published
|
|
1TEH
 
 | |
1TEI
 
 | | STRUCTURE OF CONCANAVALIN A COMPLEXED TO BETA-D-GLCNAC (1,2)ALPHA-D-MAN-(1,6)[BETA-D-GLCNAC(1,2)ALPHA-D-MAN (1,6)]ALPHA-D-MAN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Moothoo, D.N. | | Deposit date: | 1997-05-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Concanavalin A distorts the beta-GlcNAc-(1-->2)-Man linkage of beta-GlcNAc-(1-->2)-alpha-Man-(1-->3)-[beta-GlcNAc-(1-->2)-alpha-Man- (1-->6)]-Man upon binding.
Glycobiology, 8, 1998
|
|
1TEJ
 
 | | Crystal structure of a disintegrin heterodimer at 1.9 A resolution. | | Descriptor: | disintegrin chain A, disintegrin chain B | | Authors: | Bilgrami, S, Kaur, P, Yadav, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Disintegrin Heterodimer from Saw-Scaled Viper (Echis carinatus) at 1.9 A Resolution
Biochemistry, 44, 2005
|
|
1TEL
 
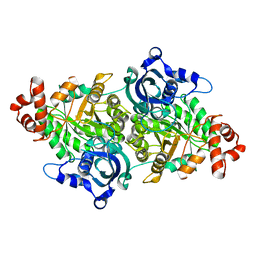 | | Crystal structure of a RubisCO-like protein from Chlorobium tepidum | | Descriptor: | ribulose bisphosphate carboxylase, large subunit | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-05-25 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a RubisCO-like protein from Chlorobium tepidum
To be Published
|
|
1TEM
 
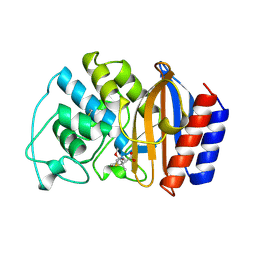 | | 6 ALPHA HYDROXYMETHYL PENICILLOIC ACID ACYLATED ON THE TEM-1 BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | 2-(1-CARBOXY-2-HYDROXY-ETHYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, TEM-1 BETA LACTAMASE | | Authors: | Maveyraud, L, Massova, I, Samama, J.P, Mobashery, S. | | Deposit date: | 1996-05-28 | | Release date: | 1997-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 6Alpha-Hydroxymethylpenicillanate Complexed to the Tem-1 Beta-Lactamase from Escherichia Coli: Evidence on the Mechanism of Action of a Novel Inhibitor Designed by a Computer-Aided Process
J.Am.Chem.Soc., 118, 1996
|
|
1TEN
 
 | |
1TEQ
 
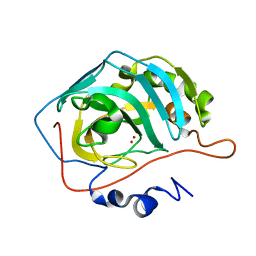 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, HYDROXIDE ION, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-25 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1TER
 
 | |
1TES
 
 | | OXYGEN BINDING MUSCLE PROTEIN | | Descriptor: | METHYLETHYLAMINE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Phillips Jr, G.N, Olson, J.S. | | Deposit date: | 1996-05-06 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of NO-induced oxidation of myoglobin and hemoglobin.
Biochemistry, 35, 1996
|
|
1TET
 
 | |
1TEU
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-25 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1TEV
 
 | | Crystal structure of the human UMP/CMP kinase in open conformation | | Descriptor: | SULFATE ION, UMP-CMP kinase | | Authors: | Segura-Pena, D, Sekulic, N, Ort, S, Konrad, M, Lavie, A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate-induced Conformational Changes in Human UMP/CMP Kinase.
J.Biol.Chem., 279, 2004
|
|
1TEW
 
 | |
1TEX
 
 | | Mycobacterium smegmatis Stf0 Sulfotransferase with Trehalose | | Descriptor: | Stf0 Sulfotransferase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Mougous, J.D, Petzold, C.J, Senaratne, R.H, Lee, D.H, Akey, D.L, Lin, F.L, Munchel, S.E, Pratt, M.R, Riley, L.W, Leary, J.A, Berger, J.M, Bertozzi, C.R. | | Deposit date: | 2004-05-25 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification, function and structure of the mycobacterial sulfotransferase that initiates sulfolipid-1 biosynthesis.
Nat.Struct.Mol.Biol., 11, 2004
|
|
