1MSG
 
 | |
1PGX
 
 | |
1RCH
 
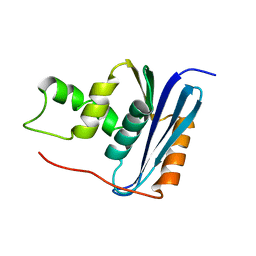 | | SOLUTION NMR STRUCTURE OF RIBONUCLEASE HI FROM ESCHERICHIA COLI, 8 STRUCTURES | | Descriptor: | RIBONUCLEASE HI | | Authors: | Yamazaki, T, Fujiwara, M, Kato, T, Yamasaki, K, Nagayama, K. | | Deposit date: | 1995-06-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribonuclease Hi from Escherichia Coli
Biol.Pharm.Bull., 23, 2000
|
|
1QCK
 
 | |
1GXG
 
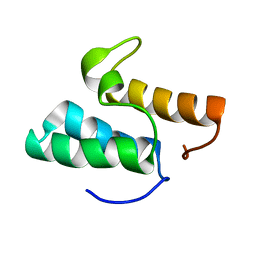 | | Non-cognate protein-protein interactions: the NMR structure of the colicin E8 inhibitor protein Im8 and its interaction with the DNase domain of colicin E9 | | Descriptor: | COLICIN E8 IMMUNITY PROTEIN | | Authors: | Le Duff, C.S, Videler, H, Boetzel, R, Czisch, M, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Non-Cognate Protein-Protein Interaction: The NMR Structure of the Colicin E8 Inhibitor Protein Im8 and its Interaction with the DNase Domain of Colicin E9
To be Published
|
|
1PFS
 
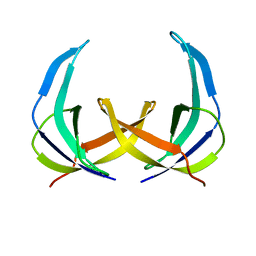 | | SOLUTION NMR STRUCTURE OF THE SINGLE-STRANDED DNA BINDING PROTEIN OF THE FILAMENTOUS PSEUDOMONAS PHAGE PF3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PF3 SINGLE-STRANDED DNA BINDING PROTEIN | | Authors: | Folmer, R.H.A, Nilges, M, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1996-08-03 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the single-stranded DNA binding protein of the filamentous Pseudomonas phage Pf3: similarity to other proteins binding to single-stranded nucleic acids.
EMBO J., 14, 1995
|
|
1ATY
 
 | |
229D
 
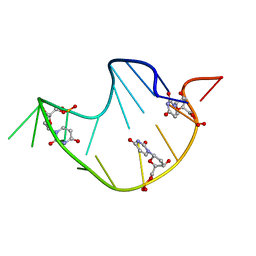 | | DNA ANALOG OF YEAST TRANSFER RNA PHE ANTICODON DOMAIN WITH MODIFIED BASES 5-METHYL CYTOSINE AND 1-METHYL GUANINE | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*CP*(UMP)P*GP*AP*AP*(MG1)P*AP*(UMP)P*(5CM)P*(UMP)P*GP*G)-3') | | Authors: | Basti, M.M, Stuart, J.W, Lam, A.T, Guenther, R, Agris, P.F. | | Deposit date: | 1995-08-16 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Design, biological activity and NMR-solution structure of a DNA analogue of yeast tRNA(Phe) anticodon domain.
Nat.Struct.Biol., 3, 1996
|
|
2RRL
 
 | | Solution structure of the C-terminal domain of the FliK | | Descriptor: | Flagellar hook-length control protein | | Authors: | Mizuno, S, Tate, S, Kobayashi, N, Amida, H. | | Deposit date: | 2011-01-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of FliK, the trigger for the switch of substrate specificity in the flagellar type III secretion apparatus
J.Mol.Biol., 409, 2011
|
|
1QR5
 
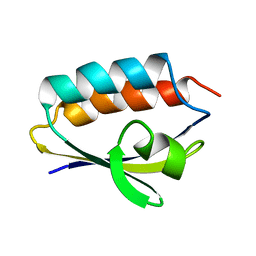 | | SOLUTION STRUCTURE OF HISTIDINE CONTAINING PROTEIN (HPR) FROM STAPHYLOCOCCUS CARNOSUS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Kalbitzer, H.R, Gorler, A, Li, H, Dubovskii, P.V, Hengstenberg, W, Kowolik, C, Yamada, H, Akasaka, K. | | Deposit date: | 1999-05-19 | | Release date: | 2000-06-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | 15N and 1H NMR study of histidine containing protein (HPr) from Staphylococcus carnosus at high pressure.
Protein Sci., 9, 2000
|
|
1HG9
 
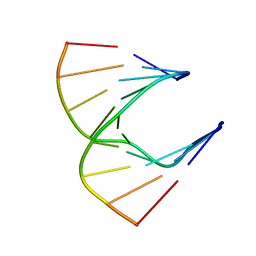 | | Solution structure of DNA:RNA hybrid | | Descriptor: | 5- D(*CP*TP*GP*AP*TP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-13 | | Release date: | 2002-01-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity.
Chemistry, 6, 2000
|
|
1HHW
 
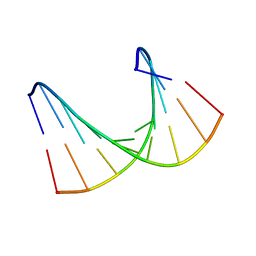 | | Solution structure of LNA1:RNA hybrid | | Descriptor: | 5- D(*CP*TP*GP*AP*+TLNP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-29 | | Release date: | 2002-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids
J.Am.Chem.Soc., 124, 2002
|
|
1MII
 
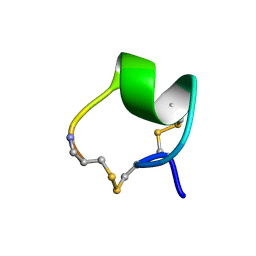 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN MII | | Descriptor: | PROTEIN (ALPHA CONOTOXIN MII) | | Authors: | Hill, J.M, Oomen, C.J, Miranda, L.P, Bingham, J.P, Alewood, P.F, Craik, D.J. | | Deposit date: | 1998-10-05 | | Release date: | 1998-10-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of alpha-conotoxin MII by NMR spectroscopy: effects of solution environment on helicity.
Biochemistry, 37, 1998
|
|
1HIC
 
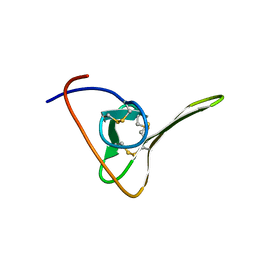 | |
1GD3
 
 | | refined solution structure of human cystatin A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix
conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
1IEZ
 
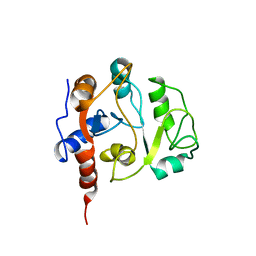 | | Solution Structure of 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase of Riboflavin Biosynthesis | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase | | Authors: | Kelly, M.J.S, Ball, L.J, Kuhne, R, Bacher, A, Oschkinat, H. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the 47-kDa dimeric enzyme 3,4-dihydroxy-2-butanone-4-phosphate synthase and ligand binding studies reveal the location of the active site.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HHX
 
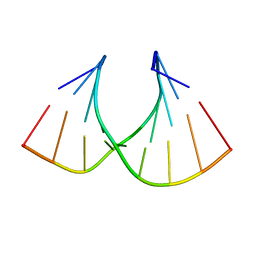 | | Solution structure of LNA3:RNA hybrid | | Descriptor: | 5- D(*CP*+TP*GP*AP*+TP*AP*+TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-29 | | Release date: | 2002-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids
J.Am.Chem.Soc., 124, 2002
|
|
1CLB
 
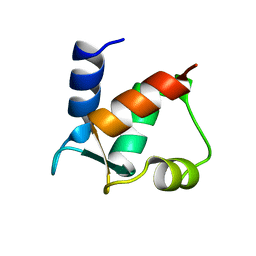 | |
1FJC
 
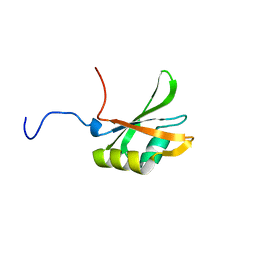 | |
1HDN
 
 | |
1FJ7
 
 | |
1GD4
 
 | | SOLUTION STRUCTURE OF P25S CYSTATIN A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
1GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
2BBG
 
 | |
1GIW
 
 | | SOLUTION STRUCTURE OF REDUCED HORSE HEART CYTOCHROME C, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Huber, J.G, Spyroulias, G.A, Turano, P. | | Deposit date: | 1998-06-17 | | Release date: | 1998-12-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced horse heart cytochrome c.
J.Biol.Inorg.Chem., 4, 1999
|
|
